

Xinjiang Agricultural Sciences ›› 2025, Vol. 62 ›› Issue (4): 800-806.DOI: 10.6048/j.issn.1001-4330.2025.04.003
• Crop Genetics and Breeding · Cultivation Physiology · Physilolgy and Biochemistry • Previous Articles Next Articles
LI Huqing( ), SHAO Dongnan, ZHANG Yi, LIU Feng, ZHANG Xinyu, LI Yanjun, SUN Jie, YANG Yonglin, XUE Fei(
), SHAO Dongnan, ZHANG Yi, LIU Feng, ZHANG Xinyu, LI Yanjun, SUN Jie, YANG Yonglin, XUE Fei( )
)
Received:2024-09-11
Online:2025-04-20
Published:2025-06-20
Supported by:
李虎情( ), 邵冬南, 张翼, 刘峰, 张新宇, 李艳军, 孙杰, 杨永林, 薛飞(
), 邵冬南, 张翼, 刘峰, 张新宇, 李艳军, 孙杰, 杨永林, 薛飞( )
)
通讯作者:
薛飞(1983-), 男,山东人,教授,博士,硕士生/博士生导师,研究方向为棉花生物育种,(E-mail)xuefei2013@hotmail.com
作者简介:李虎情(1996-),男,硕士,研究方向为棉花分子育种,(E-mail)2418645864@qq.com
基金资助:CLC Number:
LI Huqing, SHAO Dongnan, ZHANG Yi, LIU Feng, ZHANG Xinyu, LI Yanjun, SUN Jie, YANG Yonglin, XUE Fei. Bioinformatics analysis and functional verification of GHSHA1 gene in Gossypium hirsutum L.[J]. Xinjiang Agricultural Sciences, 2025, 62(4): 800-806.
李虎情, 邵冬南, 张翼, 刘峰, 张新宇, 李艳军, 孙杰, 杨永林, 薛飞. 陆地棉GhSHA1基因的特征分析及其功能验证[J]. 新疆农业科学, 2025, 62(4): 800-806.
Add to citation manager EndNote|Ris|BibTeX
URL: https://www.xjnykx.com/EN/10.6048/j.issn.1001-4330.2025.04.003
| 引物名称 Primer name | 引物序列(5’ -3’) Primer sequence(5’ -3’) | 用途 Application |
|---|---|---|
| GhSHA1 | F: GAATTCTTTTGGGCATCACTCCTCCC | VIGS |
| R: GGTACCTCACCACTGGCAGTCCTTTC | ||
| GhSHA1 | F:TCTGTGGAAATCCTGCAAAAAC | qRT-PCR |
| R:TCACTTAGTCGTTGCCTCCAAA | ||
| AtActin | F:GGTAACATTGTGCTCAGTGGTGG | qRT-PCR(内参) |
| R:AACGACCTTAATCTTCATGCTGC | ||
| GhUBQ7 | F:GAAGGCATTCCACCTGACCAAC | qRT-PCR(内参) |
| R:CTTGACCTTCTTCTTCTTGTGCTTG | ||
| AtCLV1 | F:CATCGCCCCAGAGTATGCAT | qRT-PCR |
| R:CAGTCAACCTCGGGTCAACA | ||
| AtCLV2 | F: TCCAACGCATCAAGCTCGGG | qRT-PCR |
| R:ACGCCTCTCGTCCGTCTCTT | ||
| AtCLV3 | F:TGTCCGGTCCAGTTCAACAA | qRT-PCR |
| R:CTCCCGAAATGGTAAAACCG | ||
| AtWUS | F:ACAAGCCATATCCCAGCTTCA | qRT-PCR |
| R:CCACCGTTGATGTGATCTTCA |
Tab.1 Primer sequence
| 引物名称 Primer name | 引物序列(5’ -3’) Primer sequence(5’ -3’) | 用途 Application |
|---|---|---|
| GhSHA1 | F: GAATTCTTTTGGGCATCACTCCTCCC | VIGS |
| R: GGTACCTCACCACTGGCAGTCCTTTC | ||
| GhSHA1 | F:TCTGTGGAAATCCTGCAAAAAC | qRT-PCR |
| R:TCACTTAGTCGTTGCCTCCAAA | ||
| AtActin | F:GGTAACATTGTGCTCAGTGGTGG | qRT-PCR(内参) |
| R:AACGACCTTAATCTTCATGCTGC | ||
| GhUBQ7 | F:GAAGGCATTCCACCTGACCAAC | qRT-PCR(内参) |
| R:CTTGACCTTCTTCTTCTTGTGCTTG | ||
| AtCLV1 | F:CATCGCCCCAGAGTATGCAT | qRT-PCR |
| R:CAGTCAACCTCGGGTCAACA | ||
| AtCLV2 | F: TCCAACGCATCAAGCTCGGG | qRT-PCR |
| R:ACGCCTCTCGTCCGTCTCTT | ||
| AtCLV3 | F:TGTCCGGTCCAGTTCAACAA | qRT-PCR |
| R:CTCCCGAAATGGTAAAACCG | ||
| AtWUS | F:ACAAGCCATATCCCAGCTTCA | qRT-PCR |
| R:CCACCGTTGATGTGATCTTCA |
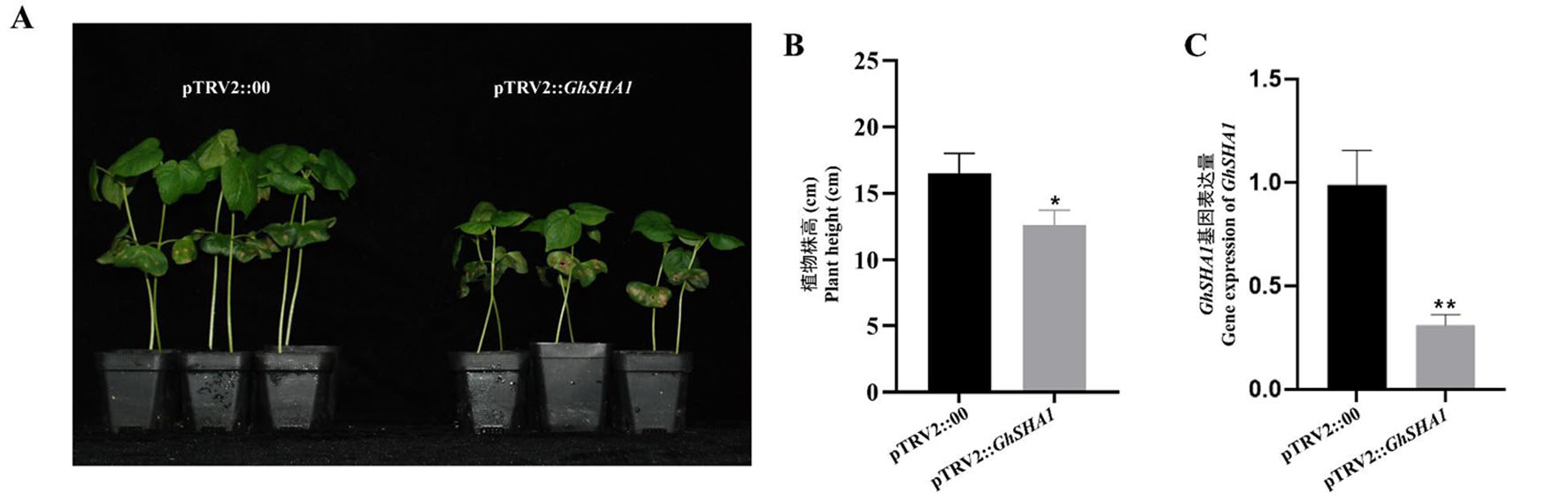
Fig.2 Changes in cotton plant height under bene GhSHA1 silencing Note:A、GhSHA1 gene VIGS Plant height;BpTRV2::GhSHA1 Chantes in Plant height after gene silencing;C、pTRV2::GhSHA1 gene silencing expression efficiency
| [1] | Wu H H, Ren Z Y, Zheng L, et al. The bHLH transcription factor GhPAS1 mediates BR signaling to regulate plant development and architecture in cotton[J]. The Crop Journal, 2021, 9(5): 1049-1059. |
| [2] |
Narnoliya L, Basu U, Bajaj D, et al. Transcriptional signatures modulating shoot apical meristem morphometric and plant architectural traits enhance yield and productivity in chickpea[J]. Plant Journal, 2019, 98(5): 864-883.
DOI |
| [3] | Xue Z H, Liu L Y, Zhang C. Regulation of shoot apical meristem and axillary meristem development in plants[J]. International Journal of Molecular Sciences, 2020, 21(8): 2917. |
| [4] | Nikolaev S V, Penenko A V, Lavreha V V, et al. A model study of the role of proteins CLV1, CLV2, CLV3, and WUS in regulation of the structure of the shoot apical meristem[J]. Russian Journal of Developmental Biology, 2007, 38(6): 383-388. |
| [5] |
Kyozuka J. Control of shoot and root meristem function by cytokinin[J]. Current Opinion in Plant Biology, 2007, 10(5): 442-446.
DOI PMID |
| [6] |
Azizi P, Rafii M Y, Maziah M, et al. Understanding the shoot apical meristem regulation: a study of the phytohormones, auxin and cytokinin, in rice[J]. Mechanisms of Development, 2015, 135: 1-15.
DOI PMID |
| [7] |
Gordon S P, Chickarmane V S, Ohno C, et al. Multiple feedback loops through cytokinin signaling control stem cell number within the Arabidopsis shoot meristem[J]. Proceedings of the National Academy of Sciences of the United States of America, 2009, 106(38): 16529-16534.
DOI PMID |
| [8] | Nidhi S, Preciado J, Tie L. Knox homologs shoot meristemless (STM) and KNAT6 are epistatic to CLAVATA3 (CLV3) during shoot meristem development in Arabidopsis thaliana[J]. Molecular Biology Reports, 2021, 48(9): 6291-6302. |
| [9] | Byrne M E, Barley R, Curtis M, et al. Asymmetric leaves1 mediates leaf patterning and stem cell function in Arabidopsis[J]. Nature, 2000, 408(6815): 967-971. |
| [10] | Scofield S, Dewitte W, Murray J A. STM sustains stem cell function in the Arabidopsis shoot apical meristem and controls KNOX gene expression independently of the transcriptional repressor AS1[J]. Plant Signaling & Behavior, 2014, 9: e28934. |
| [11] | Sonoda Y, Yao S G, Sako K, et al. SHA1, a novel RING finger protein, functions in shoot apical meristem maintenance in Arabidopsis[J]. The Plant Journal, 2007, 50(4): 586-596. |
| [12] | 易艳萍, 曹诚, 马清钧. 泛素化和磷酸化协同作用调控蛋白质降解[J]. 生物技术通讯, 2006, 17(4): 618-620. |
| YI Yanping, CAO Cheng, MA Qingjun. Ubiquitination and phosphorylation cooperate to regulate the degradation of protein[J]. Letters in Biotechnology, 2006, 17(4): 618-620. | |
| [13] |
张翼, 邵冬南, 薛飞, 等. 陆地棉细胞色素P450超家族基因GhCYP85A2-1的克隆与功能分析[J]. 新疆农业科学, 2021, 58(2): 197-205.
DOI |
|
ZHANG Yi, SHAO Dongnan, XUE Fei, et al. Cloning and functional analysis of cytochrome P450 superfamily gene GhCYP85A2-1 in upland cotton[J]. Xinjiang Agricultural Sciences, 2021, 58(2): 197-205.
DOI |
|
| [14] | Delk N A, Johnson K A, Chowdhury N I, et al. CML24, regulated in expression by diverse stimuli, encodes a potential Ca2+ sensor that functions in responses to abscisic acid, daylength, and ion stress[J]. Plant Physiology, 2005, 139(1): 240-253. |
| [15] | Tsai Y C, Delk N A, Chowdhury N I, et al. Arabidopsis potential calcium sensors regulate nitric oxide levels and the transition to flowering[J]. Plant Signaling & Behavior, 2007, 2(6): 446-454. |
| [16] | Ayadi M, Delaporte V, Li Y F, et al. Analysis of GT-3a identifies a distinct subgroup of trihelix DNA-binding transcription factors in Arabidopsis[J]. FEBS Letters, 2004, 562(1/2/3): 147-154. |
| [17] | Lee Y K, Kumari S, Olson A, et al. Role of a ZF-HD Transcription factor in miR157-Mediated feed-forward regulatory module that determines plant architecture inArabidopsis[J]. Internation Journal of Molecular Sciences, 2022, 23(15). |
| [18] |
Nikolaev SV, Penenko AV, Lavrekha VV, et al. A model study of the role of proteins CLV1, CLV2, CLV3, and WUS in regulation of the structure of the shoot apical meristem[J]. Ontogenez, 2007, 38(6):457-62.
PMID |
| [19] | Fletcher J C. The CLV-WUS stem cell signaling pathway: a roadmap to crop yield optimization[J]. Plants (Basel), 2018, 7(4): 87. |
| [20] |
Guo Y. F, Han L H, Hymes M, et al. CLAVATA2 forms a distinct CLE-binding receptor complex regulating Arabidopsis stem cell specification[J]. The Plant Journal, 2010, 63(6): 889-900.
DOI PMID |
| [21] |
XIN W, WANG Z C, LIANG Y, et al. Dynamic expression reveals a two-step patterning of WUS and CLV3 during axillary shoot meristem formation in Arabidopsis[J]. Journal of Plant Physiology, 2017, 214: 1-6.
DOI PMID |
| [22] | 高丽. 拟南芥CLAVATA3受体的研究进展[J]. 河北师范大学学报(自然科学版), 2014, 38(4): 425-432. |
| GAO Li. Research progress of CLAVATA3 receptors of Arabidopsis thaliana[J]. Journal of Hebei Normal University (Natural Science Edition), 2014, 38(4): 425-432. | |
| [23] | Durbak A R, Tax F E. CLAVATA signaling pathway receptors of Arabidopsis regulate cell proliferation in fruit organ formation as well as in meristems[J]. Genetics, 2011, 189(1): 177-194. |
| [24] | Su Y H, Zhou C, Li Y J, et al. Integration of pluripotency pathways regulates stem cell maintenance in theArabidopsisshoot meristem[J]. Proceedings of the National Academy of Sciences of the United States of America, 2020, 117(36): 22561-22571. |
| [25] | Diévart A, Dalal M, Tax F E, et al. CLAVATA1 dominant-negative alleles reveal functional overlap between multiple receptor kinases that regulate meristem and organ development[J]. The Plant Cell, 2003, 15(5): 1198-1211. |
| [1] | BAO Yanli, WANG Xiaowei, LI Qiongshi, ZHANG Lizhao, CHEN Yulan. Analysis of the high Quality development level and differences of cotton in major cotton regions of Xinjiang [J]. Xinjiang Agricultural Sciences, 2025, 62(4): 1032-1040. |
| [2] | ZHAO Yuhang, YAN An, MA Mengqian, XIAO Shuting, SUN Zhe, LI Jingyan. Estimation of cotton LAI and SPAD under water-nitrogen coupling based on multi-spectral imaging of unmanned aerial vehicle [J]. Xinjiang Agricultural Sciences, 2025, 62(4): 781-790. |
| [3] | LI Ke, YIN Caixia, CHEN Maoguang, CUI Hanyu, WANG Ke, LIU Liyang, TANG Qiuxiang. Research on cotton SPAD estimation based on UAV multispectral images combined with machine learning [J]. Xinjiang Agricultural Sciences, 2025, 62(4): 791-799. |
| [4] | QIAO Di, LIN Tao, CUI Jianping, ZHANG Pengzhong, ZHANG Hao, BAO Longlong, TANG Qiuxiang. Effects of RZWQM2-based nitrogen fertilizer transport mode on cotton growth and yield [J]. Xinjiang Agricultural Sciences, 2025, 62(4): 807-819. |
| [5] | ZHANG Lingjian, ZHANG Kai, ZHANG Hui, GUO Xiaomeng, CHEN Guoyue, WANG Yiding, JIA Qingyu. Study on the relationship between plant water content and morphological characteristics of top stem and leaf during the whole growth period of cotton [J]. Xinjiang Agricultural Sciences, 2025, 62(3): 531-538. |
| [6] | ZHANG Lian, CHEN Xiangyao, WANG Tangang, MA Xiaomei, CHENG Bin, WANG Gang, DUAN Zhenyu. Effects of high-strength mulch on soil temperature, humidity and cotton growth [J]. Xinjiang Agricultural Sciences, 2025, 62(3): 539-545. |
| [7] | XU Shouzhen, MA Qi, NING Xinzhu, LI Jilian, SU Junji, HAN Huanyong, WANG Fangyong, LIN Hai. Effects of different row spacing and defoliant on cotton defoliation [J]. Xinjiang Agricultural Sciences, 2025, 62(3): 546-555. |
| [8] | ZHAO Yupeng, CHEN Bolang, WANG Zhiguo, FU Yanbo, BIAN Qingyong. Effects of different carbon source inputs on the characteristics of compacted clay and the growth of cotton seedlings [J]. Xinjiang Agricultural Sciences, 2025, 62(3): 556-571. |
| [9] | WANG Yongpan, MA Jun, LI Chenyu, YAO Mengyao, WANG Zixuan, HUANG Lingzhi, ZHU Haiyan, LIU Wanrong, LI Bo, YANG Yang, GAO Wenwei. Salt tolerance in germination period of cotton seeds based on convolutional neural network and synthetic dataset [J]. Xinjiang Agricultural Sciences, 2025, 62(2): 261-269. |
| [10] | HU Shasha, SHAO Liping, CHEN Lihua, SONG Weiping, ZHAO Hai, ZHANG Xinyu, SUN Jie. Effect of defoliant on boll development and fiber quality of machine-picked cotton [J]. Xinjiang Agricultural Sciences, 2025, 62(2): 270-277. |
| [11] | LU Mingkun, LI Junhong, Nilupaier Yusufujiang, PAN Xipeng, LIU Xiaocheng, ZHANG Zhenggui, PAN Zhanlei, ZHAI Menghua, ZHANG Yaopeng, ZHAO Wenqi, WANG Lihong, WANG Zhanbiao. Effect of silic on fertiliser application on the growth and development of cotton and its yield and quality [J]. Xinjiang Agricultural Sciences, 2025, 62(2): 286-293. |
| [12] | WANG Yiding, ZHANG Kai, ZHANG Lingjian, ZHANG Hui, GUO Xiaomeng, CHEN Guoyue. Effects of drip irrigation on the growth and development, yield formation, and water use efficiency of cotton in Xinjiang [J]. Xinjiang Agricultural Sciences, 2025, 62(2): 294-301. |
| [13] | WANG Xiaoyan, BAI Yungang, CHAI Zhongping, LU Zhenlin, LIU Hongbo, XIAO Jun, Amannisa . Effect of "dry sowing and wet emergence" on cotton growth and yield under the control of winter drip irrigation in off-cropping period [J]. Xinjiang Agricultural Sciences, 2025, 62(2): 302-313. |
| [14] | CHENG Guanfu, LU Guoqiang, CUI Yongxiang, HOU Aocheng, ZHANG Guoshuai, LIANG Chunyan, LEI Jianfeng, LU Wei, DAI Peihong, LI yue. Functional analysis of a C2H2 zinc protein GhZFP8 gene in Gossypium hirsutum resistance to Verticillium wilt [J]. Xinjiang Agricultural Sciences, 2025, 62(1): 137-145. |
| [15] | SUN Caiqin, WU Jia, HUANG Hai, GUO Jiaxin, MIN Wei, GUO Huijuan. Effects of different saline and alkaline stress on the proteome of cotton root system [J]. Xinjiang Agricultural Sciences, 2025, 62(1): 146-160. |
| Viewed | ||||||
|
Full text |
|
|||||
|
Abstract |
|
|||||