

Xinjiang Agricultural Sciences ›› 2025, Vol. 62 ›› Issue (1): 137-145.DOI: 10.6048/j.issn.1001-4330.2025.01.017
• Cultivation Physiology·Physiology and Biochemistry·Germplasm Resources·Molecular Genetics·Soil Fertilizer • Previous Articles Next Articles
CHENG Guanfu( ), LU Guoqiang, CUI Yongxiang, HOU Aocheng, ZHANG Guoshuai, LIANG Chunyan, LEI Jianfeng, LU Wei, DAI Peihong, LI yue(
), LU Guoqiang, CUI Yongxiang, HOU Aocheng, ZHANG Guoshuai, LIANG Chunyan, LEI Jianfeng, LU Wei, DAI Peihong, LI yue( )
)
Received:2024-08-11
Online:2025-01-20
Published:2025-03-11
Correspondence author:
LI yue
Supported by:
程贯富( ), 卢国强, 崔永祥, 侯奥成, 张国帅, 梁春燕, 雷建峰, 路伟, 代培红, 李月(
), 卢国强, 崔永祥, 侯奥成, 张国帅, 梁春燕, 雷建峰, 路伟, 代培红, 李月( )
)
通讯作者:
李月
作者简介:程贯富(1998-),男,山东济南人,硕士研究生,研究方向为棉花逆境分子生物学,(E-mail)cgfyouxi@163.com
基金资助:CLC Number:
CHENG Guanfu, LU Guoqiang, CUI Yongxiang, HOU Aocheng, ZHANG Guoshuai, LIANG Chunyan, LEI Jianfeng, LU Wei, DAI Peihong, LI yue. Functional analysis of a C2H2 zinc protein GhZFP8 gene in Gossypium hirsutum resistance to Verticillium wilt[J]. Xinjiang Agricultural Sciences, 2025, 62(1): 137-145.
程贯富, 卢国强, 崔永祥, 侯奥成, 张国帅, 梁春燕, 雷建峰, 路伟, 代培红, 李月. 陆地棉锌指蛋白基因GhZFP8黄萎病的抗性功能验证[J]. 新疆农业科学, 2025, 62(1): 137-145.
| 分析工具 Analysis tools | 功能 Function | 网址 URL |
|---|---|---|
| ExPASy-ProtScale | 蛋白理化性质分析 | |
| SignaIP | 信号肽预测 | |
| TMHMM | 跨膜结构域预测 | |
| SMART | 蛋白结构域预测 | |
| PSIPRED | 蛋白二级结构预测 | |
| SWISS-MODEL | 蛋白三级结构预测 | |
Tab.1 Bioinformatics analysis tools
| 分析工具 Analysis tools | 功能 Function | 网址 URL |
|---|---|---|
| ExPASy-ProtScale | 蛋白理化性质分析 | |
| SignaIP | 信号肽预测 | |
| TMHMM | 跨膜结构域预测 | |
| SMART | 蛋白结构域预测 | |
| PSIPRED | 蛋白二级结构预测 | |
| SWISS-MODEL | 蛋白三级结构预测 | |
| 基因 Gemne | 序列 Sequence(5'-3') | 用途 Application |
|---|---|---|
| GhZFP8 | F:TCGGAAGTATTTGATATGGCGCTTG R:GCTTTGGACCTCTTCTTCATCTGGG | 基因克隆 |
| VIGS- GhZFP8 | F: GAATTCACAACGTGGCGACTCTCCT R: GGTACCAGCGTTTATGACCACCCAAG | VIGS载体构建 |
| qGhZFP8 | F:ATTCGTCGGTCTTCCCCTTC R:TTTGCGGTGGCTGGCTTTAT | qRT-PCR |
| qGhUBQ7 | F:GAAGGCATTCCACCTGACCAAC R:CTTGACCTTCTTCTTCTTGTGCTTG | qRT-PCR |
Tab.2 Primer and application
| 基因 Gemne | 序列 Sequence(5'-3') | 用途 Application |
|---|---|---|
| GhZFP8 | F:TCGGAAGTATTTGATATGGCGCTTG R:GCTTTGGACCTCTTCTTCATCTGGG | 基因克隆 |
| VIGS- GhZFP8 | F: GAATTCACAACGTGGCGACTCTCCT R: GGTACCAGCGTTTATGACCACCCAAG | VIGS载体构建 |
| qGhZFP8 | F:ATTCGTCGGTCTTCCCCTTC R:TTTGCGGTGGCTGGCTTTAT | qRT-PCR |
| qGhUBQ7 | F:GAAGGCATTCCACCTGACCAAC R:CTTGACCTTCTTCTTCTTGTGCTTG | qRT-PCR |
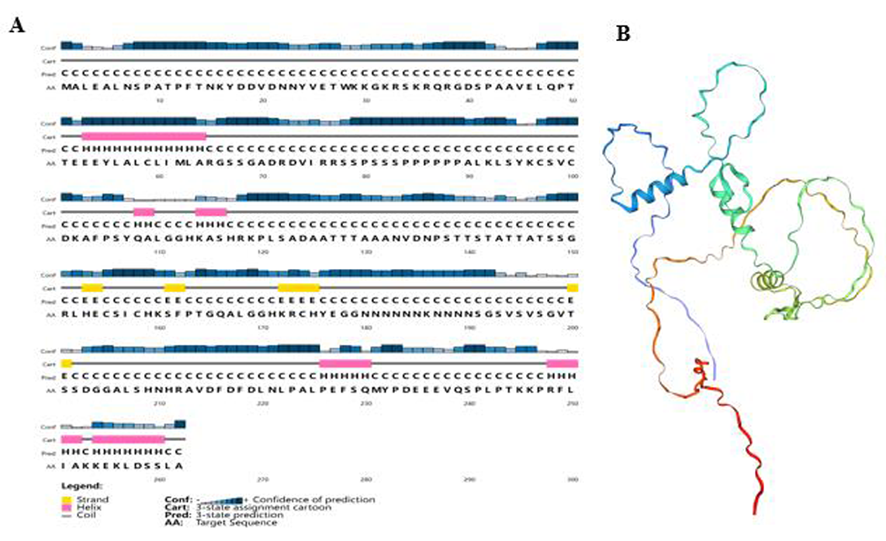
Fig.4 The prediction secondary and tertiary structure of GhZFP8 protein Notes: A: The prediction secondary structure of GhZFP8 protein; B: The prediction tertiary structure of GhZFP8protein
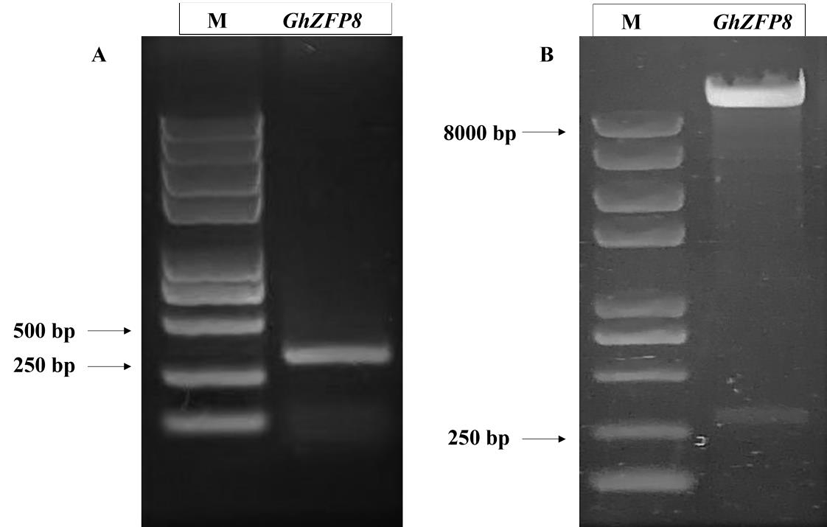
Fig.5 Construction of VIGS vector of GhZFP8 Notes: A:PCR amplification of target fragment of GhZFP8 ;B :identification of TRV: GhZFP8 VIGS vectors by restriction enzyme digestion.M :Trans 2K Plus Ⅱ DNA marker
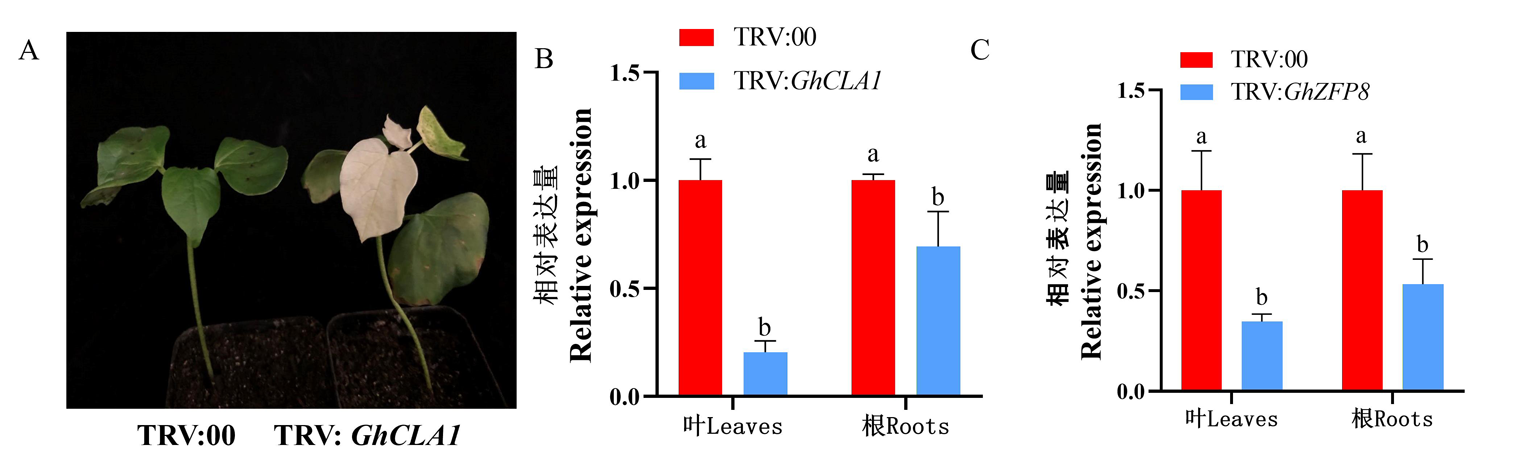
Fig.6 Detection results of silencing efficiency of GhZFP8 gene Notes:A:Phenotype of positive control plant;B:Silencing efficiency of GhCLA1 gene;C:Silencing efficiency of GhZFP8 gene; The same lowercase letter indicates no significant difference(P>0.05),and different lowercase letters indicate significant difference (P<0.05),the same as below

Fig.7 The Phenotype and disease index of silenced plants inoculated with Verticillium Notes: A: Phenotype analysis of plants after GhZFP8gene silencing and Identification by stem anatomy.B: Disease index statistics.C: the stained section of Xylem
| [1] | Li F G, Fan G Y, Lu C R, et al. Genome sequence of cultivated Upland cotton (Gossypium hirsutum TM-1) provides insights into genome evolution[J]. Nature Biotechnology, 2015, 33(5): 524-530. |
| [2] | 马存, 简桂良, 郑传临. 中国棉花抗枯、黄萎病育种50年[J]. 中国农业科学, 2002, 35(5): 508-513. |
| MA Cun, JIAN Guiliang, ZHENG Chuanlin. The advances in cotton breeding resistance to Fusarium and Verticillium wilts in China during past fifty years[J]. Scientia Agricultura Sinica, 2002, 35(5): 508-513. | |
| [3] |
Wei Z, Yu D. Analysis of the succession of structure of the bacteria community in soil from long-term continuous cotton cropping in Xinjiang using high-throughput sequencing[J]. Archives of Microbiology, 2018, 200(4): 653-662.
DOI PMID |
| [4] |
Mohamed H I, Akladious S A. Changes in antioxidants potential, secondary metabolites and plant hormones induced by different fungicides treatment in cotton plants[J]. Pesticide Biochemistry and Physiology, 2017, 142: 117-122.
DOI PMID |
| [5] |
Zhang Z B, Li X L, Yu R, et al. Isolation, structural analysis, and expression characteristics of the maize TIFY gene family[J]. Molecular Genetics and Genomics, 2015, 290(5): 1849-1858.
DOI PMID |
| [6] | Yu Y H, Wan Y T, Jiao Z L, et al. Functional characterization of resistance to powdery mildew of VvTIFY9 from Vitis vinifera[J]. International Journal of Molecular Sciences, 2019, 20(17): 4286. |
| [7] | Zang Z Y, Lv Y, Liu S, et al. A novel ERF transcription factor, ZmERF105, positively regulates maize resistance to Exserohilum turcicum[J]. Frontiers in Plant Science, 2020, 11: 850. |
| [8] | Liu Y H, Khan A R, Gan Y B. C2H2 zinc finger proteins response to abiotic stress in plants[J]. International Journal of Molecular Sciences, 2022, 23(5): 2730. |
| [9] |
Berg J M, Shi Y. The galvanization of biology: a growing appreciation for the roles of zinc[J]. Science, 1996, 271(5252): 1081-1085.
DOI PMID |
| [10] | Zheng F Y, Cui L, Li C X, et al. Hair interacts with SlZFP8-like to regulate the initiation and elongation of trichomes by modulating SlZFP6 expression in tomato[J]. Journal of Experimental Botany, 2022, 73(1): 228-244. |
| [11] | Mittler R, Kim Y, Song L H, et al. Gain- and loss-of-function mutations in Zat10 enhance the tolerance of plants to abiotic stress[J]. FEBS Letters, 2006, 580(28/29): 6537-6542. |
| [12] |
Davletova S, Schlauch K, Coutu J, et al. The zinc-finger protein Zat12 plays a central role in reactive oxygen and abiotic stress signaling in Arabidopsis[J]. Plant Physiology, 2005, 139(2): 847-856.
DOI PMID |
| [13] |
Kim J C, Jeong J C, Park H C, et al. Cold accumulation of SCOF-1 transcripts is associated with transcriptional activation and mRNA stability[J]. Molecules and Cells, 2001, 12(2): 204-208.
PMID |
| [14] |
Tian Z D, Zhang Y, Liu J, et al. Novel potato C2H2-type zinc finger protein gene, StZFP1, which responds to biotic and abiotic stress, plays a role in salt tolerance[J]. Plant Biology, 2010, 12(5): 689-697.
DOI PMID |
| [15] |
Uehara Y, Takahashi Y, Berberich T, et al. Tobacco ZFT1, a transcriptional repressor with a Cys2/His2 type zinc finger motif that functions in spermine-signaling pathway[J]. Plant Molecular Biology, 2005, 59(3): 435-448.
DOI PMID |
| [16] |
Li W T, Zhu Z W, Chern M, et al. A natural allele of a transcription factor in rice confers broad-spectrum blast resistance[J]. Cell, 2017, 170(1): 114-126.
DOI PMID |
| [17] |
王亚, 王越涛, 申关望, 等. 聚合R基因Pigm和非R基因bsr-d1改良水稻稻瘟病抗性[J]. 华北农学报, 2022, 37(5): 157-165.
DOI |
| WANG Ya, WANG Yuetao, SHEN Guanwang, et al. Improvement of rice blast resistance by pyramiding the R gene pigm and the non-R gene bsr-d1[J]. Acta Agriculturae Boreali-Sinica, 2022, 37(5): 157-165. | |
| [18] | Gu Z H, Liu T L, Ding B, et al. Two lysin-motif receptor kinases, gh-LYK1 and gh-LYK2, contribute to resistance against Verticillium wilt in upland cotton[J]. Frontiers in Plant Science, 2017, 8: 2133. |
| [19] | Wang G L, Xu J, Li L C, et al. GbCYP86A1-1 from Gossypium barbadense positively regulates defence against Verticillium dahliae by cell wall modification and activation of immune pathways[J]. Plant Biotechnology Journal, 2020, 18(1): 222-238. |
| [20] |
Wang P, Zhou L, Jamieson P, et al. The cotton wall-associated kinase GhWAK7A mediates responses to fungal wilt pathogens by complexing with the chitin sensory receptors[J]. The Plant Cell, 2020, 32(12): 3978-4001.
DOI PMID |
| [21] | Feng H J, Li C, Zhou J L, et al. A cotton WAKL protein interacted with a DnaJ protein and was involved in defense against Verticillium dahliae[J]. International Journal of Biological Macromolecules, 2021, 167: 633-643. |
| [22] | 胡子曜, 武晓玉, 雷建峰, 等. 陆地棉小GTP结合蛋白基因GhROP1和GhROP8的克隆及表达分析[J]. 中国农业大学学报, 2023, 28(4): 13-25. |
| HU Ziyao, WU Xiaoyu, LEI Jianfeng, et al. Molecular cloning and expression analysis of small GTP-binding protein genes GhROP1 and GhROP8 in cotton (Gossypium hirsutum L.)[J]. Journal of China Agricultural University, 2023, 28(4): 13-25. | |
| [23] | 李秀青, 李月, 刘超, 等. 棉花黄萎病相关基因GhAAT的克隆与功能鉴定[J]. 分子植物育种, 2020, 18(4): 1048-1053. |
| LI Xiuqing, LI Yue, LIU Chao, et al. Cloning and functional identification of cotton Verticillium wilt related gene GhAAT[J]. Molecular Plant Breeding, 2020, 18(4): 1048-1053. | |
| [24] | Hou T Z, Huang M Z, Liao Y, et al. Virus-induced gene silencing (VIGS) for functional analysis of genes involved in the regulation of anthocyanin biosynthesis in the perianth of Phalaenopsis-type Dendrobium hybrids[J]. Scientia Horticulturae, 2023, 307: 111485. |
| [25] |
袁伟, 万红建, 杨悦俭. 植物实时荧光定量PCR内参基因的特点及选择[J]. 植物学报, 2012, 47(4): 427-436.
DOI |
| YUAN Wei, WAN Hongjian, YANG Yuejian. Characterization and selection of reference genes for real-time quantitative RT-PCR of plants[J]. Chinese Bulletin of Botany, 2012, 47(4): 427-436. | |
| [26] | 王倩, 章超, 代培红, 等. 陆地棉GhMAPKKKK3基因的克隆及其表达分析[J]. 分子植物育种, 2024, 22(3): 697-703. |
| WANG Qian, ZHANG Chao, DAI Peihong, et al. Cloning and expression analysis of GhMAPKKKK3 gene in Gossypium hirsutum L[J]. Molecular Plant Breeding, 2024, 22(3): 697-703. | |
| [27] |
Livak K J, Schmittgen T D. Analysis of relative gene expression data using real-time quantitative PCR and the 2-ΔΔC T method[J]. Methods, 2001, 25(4): 402-408.
DOI PMID |
| [28] | Li Y, Zhou Y J, Dai P H, et al. Cotton Bsr-k1 modulates lignin deposition participating in plant resistance against Verticillium dahliae and Fusarium oxysporum[J]. Plant Growth Regulation, 2021, 95(2): 283-292. |
| [29] | Dong Q, Magwanga R O, Cai X Y, et al. Rna-sequencing, physiological and RNAi analyses provide insights into the response mechanism of the ABC-mediated resistance to Verticillium dahliae infection in cotton[J]. Genes, 2019, 10(2): 110. |
| [30] | Qin T, Liu S M, Zhang Z N, et al. GhCyP3 improves the resistance of cotton to Verticillium dahliae by inhibiting the E3 ubiquitin ligase activity of GhPUB17[J]. Plant Molecular Biology, 2019, 99(4/5): 379-393. |
| [31] |
Han L B, Li Y B, Wang F X, et al. The cotton apoplastic protein CRR1 stabilizes chitinase 28 to facilitate defense against the fungal pathogen Verticillium dahliae[J]. The Plant Cell, 2019, 31(2): 520-536.
DOI PMID |
| [32] | Gong Q, Yang Z E, Chen E Y, et al. A phi-class glutathione S-transferase gene for Verticillium wilt resistance in Gossypium arboreum identified in a genome-wide association study[J]. Plant & Cell Physiology, 2018, 59(2): 275-289. |
| [33] | He X, Wang T Y, Zhu W, et al. GhHB12, a HD-ZIP I transcription factor, negatively regulates the cotton resistance to Verticillium dahliae[J]. International Journal of Molecular Sciences, 2018, 19(12): 3997. |
| [34] | Zhang H J, Zhao T Y, Zhuang P T, et al. NbCZF1, a novel C2H2-type zinc finger protein, as a new regulator of SsCut-induced plant immunity in Nicotiana benthamiana[J]. Plant & Cell Physiology, 2016, 57(12): 2472-2484. |
| [35] |
Noman A, Liu Z Q, Yang S, et al. Expression and functional evaluation of CaZNF830 during pepper response to Ralstonia solanacearum or high temperature and humidity[J]. Microbial Pathogenesis, 2018, 118: 336-346.
DOI PMID |
| [36] | Shi H T, Wang X, Ye T T, et al. The Cysteine2/Histidine2-Type Transcription Factor zinc finger of Arabidopsis thaliana6 Modulates Biotic and Abiotic Stress Responses by Activating Salicylic Acid-Related Genes and c-repeat-binding factor Genes in Arabidopsis[J]. Plant Physiology, 2014, 165(3): 1367-1379. |
| [1] | LAI Chengxia, YANG Yanlong, WANG Penglong, ZHU Mengyu, YANG Dong, LI Chunping, GE Fengwei, Mayila Yusuyin, YANG Ni, MA Jun. Pathogen colony’s morphological characteristics and pathogenicity identification of defioating cotton Verticillium wilt in some areas in northern Xinjiang [J]. Xinjiang Agricultural Sciences, 2025, 62(1): 174-181. |
| [2] | LAI Chengxia, YANG Yanlong, LI Chunping, Mayila Yusuyin, WANG Yan, YANG Dong, YANG Ni, GE Fengwei, WANG Penglong, MA Jun. Biological characteristics and chemical control of defoliating cotton Verticillium wilt [J]. Xinjiang Agricultural Sciences, 2024, 61(8): 2034-2042. |
| [3] | MA Baihuan, ZHAO Qiang, XIE Jia, XU Kaiyue, REN Ruofei, SONG Xinghu. Effects of biopharmaceutical mixture on the control and growth of cotton Verticillium wilt [J]. Xinjiang Agricultural Sciences, 2024, 61(7): 1748-1756. |
| [4] | MA Shangjie, LI Shengmei, YANG Tao, WANG Honggang, ZHAO Kang, PANG Bo, GAO Wenwei. Cloning and subcellular localization of the GHWAT1-35 gene in Gossypium hirsutum [J]. Xinjiang Agricultural Sciences, 2024, 61(6): 1310-1317. |
| [5] | OUYANG Danhua, ZHAO Kang, SONG Dongbo, LIU Ziqing, GUO Wangzhen, LIU Yan, GU Aixing, Azhatiguli Maimaitituer, Alikaerjiang Amaier. Identification and comprehensive analysis of Verticillium wilt resistance in 35 cotton strains [J]. Xinjiang Agricultural Sciences, 2024, 61(1): 9-18. |
| [6] | JIN Juan, Subina Xiaokelaiti, Abudoukayoumu Ayimaiti, YANG Lei, HAO Qing, FAN Dingyu. Cloning and sequence analysis of expansin gene ZjEXPA8 in Huizao [J]. Xinjiang Agricultural Sciences, 2023, 60(9): 2223-2230. |
| [7] | YANG Ni, Mayila Yusuyin, YANG Yanlong, LI Chunping, ZHANG Dawei, XU Haijiang, LAI Chengxia. Comparative analysis of plant volatiles from the Verticillium-Infected withered spot and etiolated leaves in cotton [J]. Xinjiang Agricultural Sciences, 2023, 60(8): 1975-1986. |
| [8] | GENG Feifei, MENG Chaomin, QING Guixia, ZHOU Jiamin, ZHANG Fuhou, LIU Fengju. Cloning and expression analysis of phosphorus efficient gene GhMYB4 in Gossypium hirsutum L. [J]. Xinjiang Agricultural Sciences, 2023, 60(6): 1406-1412. |
| [9] | CHENG Lihua, YANG Honglan, MA Qingqian, SHI Ying, ZHANG Dawei, Alisher A. Abdullaev, ZHANG Daoyuan. Physiological identification and analysis of Verticillium wilt resistance of 10 foreign cotton germplasm resources [J]. Xinjiang Agricultural Sciences, 2023, 60(4): 992-1002. |
| [10] | MA Qingqian, YANG Honglan, WEI Xin, ZHANG Dawei, Alisher A Abdullaev, CHENG Lihua, ZHANG Daoyuan. Identification and Screen from Eighteen Foreign Cotton Germplasm on Agronomy and Verticillium Resistance [J]. Xinjiang Agricultural Sciences, 2023, 60(2): 286-294. |
| [11] | Nasibai Abuduwahapu, GAO Xiaojuan, Kunduziayi Abudushalamu, LI Jiangwei. Cloning and Sequence Analysis of cDNA of C3d Gene of Bactrian Camel from Xinjiang [J]. Xinjiang Agricultural Sciences, 2023, 60(2): 493-500. |
| [12] | LIU Chenxi, ZHU Yuting, ZHOU Qiang, CHEN Jin, ZHAO Wenjie, ZHENG Kai. Cloning and bioinformatics analysis of β-carotene isomerase GbD27-6 gene in sea island cotton [J]. Xinjiang Agricultural Sciences, 2023, 60(12): 2869-2877. |
| [13] | WANG Haitao, LIU Cunjing, TANG Liyuan, ZHANG Sujun, CAI Xiao, LI Xinghe, MA Wenna, HAN Junwei, ZHANG Xiangyun, ZHANG Jianhong. The influence of different planting densities on agronomic traits, yield and quality of machine-picked cotton varieties [J]. Xinjiang Agricultural Sciences, 2023, 60(11): 2638-2645. |
| [14] | LIU Haiyang, WANG Wei, ZHANG Renfu, Wenqiemu Abulizi, YAO Ju. A Brief Analysis of the Factors Restricting the Effectiveness of Controlling Cotton Verticillium wilt by Using Biocontrol Bacteria in the Field [J]. Xinjiang Agricultural Sciences, 2022, 59(1): 155-161. |
| [15] | LIU Haiyang, WANG Wei, ZHANG Renfu, Wenqiemu Abulizi, YAO Ju. Quantitative Characteristics of Microsclerotia of Verticillium dahliae in Cotton Field Soil under Different Factors [J]. Xinjiang Agricultural Sciences, 2021, 58(3): 522-531. |
| Viewed | ||||||||||||||||||||||||||||||||||||||||||||||||||
|
Full text 14
|
|
|||||||||||||||||||||||||||||||||||||||||||||||||
|
Abstract 81
|
|
|||||||||||||||||||||||||||||||||||||||||||||||||