

Xinjiang Agricultural Sciences ›› 2024, Vol. 61 ›› Issue (3): 708-718.DOI: 10.6048/j.issn.1001-4330.2024.03.021
• Plant Protection·Microbes • Previous Articles Next Articles
YAO Yuxiang1,2( ), WANG Guoqiang2, WANG Ke2, YI Sha2, YANG Xinya2, LUO Xioaxia1,2(
), WANG Guoqiang2, WANG Ke2, YI Sha2, YANG Xinya2, LUO Xioaxia1,2( )
)
Received:2023-07-15
Online:2024-03-20
Published:2024-04-19
Correspondence author:
LUO Xiaoxia(1982-),female,from Aral,Xinjiang,professor,research interests:microbial resources and their utilization,(E-mail) Supported by:
姚宇翔1,2( ), 王国强2, 王可2, 伊莎2, 杨欣雅2, 罗晓霞1,2(
), 王国强2, 王可2, 伊莎2, 杨欣雅2, 罗晓霞1,2( )
)
通讯作者:
罗晓霞(1982-),女,新疆阿拉尔人,教授,博士,硕士生导师,研究方向为微生物资源及其利用,(E-mail)作者简介:姚宇翔(1999-),女,河南周口人,硕士研究生,研究方向为微生物资源及其利用,(E-mail)1366307897@qq.com
基金资助:CLC Number:
YAO Yuxiang, WANG Guoqiang, WANG Ke, YI Sha, YANG Xinya, LUO Xioaxia. Analysis of the bacterial communities structure and diversty of the Tarim River in different habitats[J]. Xinjiang Agricultural Sciences, 2024, 61(3): 708-718.
姚宇翔, 王国强, 王可, 伊莎, 杨欣雅, 罗晓霞. 塔里木河上游不同生境细菌群落结构及多样性分析[J]. 新疆农业科学, 2024, 61(3): 708-718.
Add to citation manager EndNote|Ris|BibTeX
URL: https://www.xjnykx.com/EN/10.6048/j.issn.1001-4330.2024.03.021
| Sample | Phylum | Class | Order | Family | Genus | Species |
|---|---|---|---|---|---|---|
| ZF | 19 | 43 | 99 | 143 | 226 | 88 |
| EG | 26 | 54 | 129 | 160 | 236 | 82 |
| TA | 21 | 54 | 103 | 142 | 178 | 47 |
| BM | 22 | 44 | 105 | 156 | 221 | 70 |
| S-T | 28 | 67 | 112 | 138 | 112 | 32 |
| S-M | 25 | 60 | 89 | 101 | 78 | 13 |
| S-Y | 26 | 55 | 97 | 120 | 95 | 36 |
| S-m | 31 | 72 | 107 | 129 | 102 | 27 |
| S4 | 26 | 62 | 116 | 143 | 117 | 30 |
| S3 | 21 | 51 | 91 | 111 | 91 | 19 |
| S2 | 28 | 65 | 110 | 136 | 114 | 33 |
| S1 | 27 | 72 | 114 | 142 | 121 | 30 |
Tab.1 Statistical table of the number of microbial classification units at all levels
| Sample | Phylum | Class | Order | Family | Genus | Species |
|---|---|---|---|---|---|---|
| ZF | 19 | 43 | 99 | 143 | 226 | 88 |
| EG | 26 | 54 | 129 | 160 | 236 | 82 |
| TA | 21 | 54 | 103 | 142 | 178 | 47 |
| BM | 22 | 44 | 105 | 156 | 221 | 70 |
| S-T | 28 | 67 | 112 | 138 | 112 | 32 |
| S-M | 25 | 60 | 89 | 101 | 78 | 13 |
| S-Y | 26 | 55 | 97 | 120 | 95 | 36 |
| S-m | 31 | 72 | 107 | 129 | 102 | 27 |
| S4 | 26 | 62 | 116 | 143 | 117 | 30 |
| S3 | 21 | 51 | 91 | 111 | 91 | 19 |
| S2 | 28 | 65 | 110 | 136 | 114 | 33 |
| S1 | 27 | 72 | 114 | 142 | 121 | 30 |

Fig.1 Analysis of bacterial community structure among different samples Note:A:Histogram of horizontal species composition of different soil phylums in the upper reaches of Tarim River,B:Histogram of horizontal species composition of different phylums of the upper reaches of Tarim River,C:Horizontal species composition of different soil genus in the upper reaches of Tarim River,D:Tarim Histogram of horizontal species composition of different water sample genera in the upper reaches of the river
| Sample | Chao1 | Simpson | Shannon | Observed_species | Faith_pd | Goods_coverage |
|---|---|---|---|---|---|---|
| S1 | 3 377.93 | 0.990 888 | 9.297 34 | 3 249 | 306.966 | 0.994 019 |
| S2 | 3 922.37 | 0.997 72 | 10.315 1 | 3 782 | 320.958 | 0.992 133 |
| S3 | 2 191.48 | 0.980 822 | 8.072 1 | 2 042 | 206.968 | 0.994 716 |
| S4 | 4 757.41 | 0.995 194 | 10.298 6 | 4 583 | 375.849 | 0.989 907 |
| S-M | 3 705.76 | 0.968 635 | 8.987 97 | 3 508 | 281.87 | 0.991 298 |
| S-m | 4 886.6 | 0.997 765 | 10.578 | 4 738 | 373.577 | 0.989 555 |
| S-T | 5 273.96 | 0.996 183 | 10.872 6 | 5 260 | 400.71 | 0.996 595 |
| S-Y | 4 452.27 | 0.998 124 | 10.712 5 | 4 230 | 364.662 | 0.990 527 |
| BM | 3 900.91 | 0.884 219 | 7.023 68 | 3 830 | 180.009 | 0.995 676 |
| EG | 3 070.09 | 0.965 024 | 7.240 96 | 3 063 | 178.746 | 0.998 786 |
| TA | 912.77 | 0.978 809 | 7.514 6 | 910 | 72.96 | 0.999 898 |
| ZF | 5 244.04 | 0.957 637 | 8.139 92 | 5 195 | 210.571 | 0.995 141 |
Tab.2 Alpha diversity index of different samples
| Sample | Chao1 | Simpson | Shannon | Observed_species | Faith_pd | Goods_coverage |
|---|---|---|---|---|---|---|
| S1 | 3 377.93 | 0.990 888 | 9.297 34 | 3 249 | 306.966 | 0.994 019 |
| S2 | 3 922.37 | 0.997 72 | 10.315 1 | 3 782 | 320.958 | 0.992 133 |
| S3 | 2 191.48 | 0.980 822 | 8.072 1 | 2 042 | 206.968 | 0.994 716 |
| S4 | 4 757.41 | 0.995 194 | 10.298 6 | 4 583 | 375.849 | 0.989 907 |
| S-M | 3 705.76 | 0.968 635 | 8.987 97 | 3 508 | 281.87 | 0.991 298 |
| S-m | 4 886.6 | 0.997 765 | 10.578 | 4 738 | 373.577 | 0.989 555 |
| S-T | 5 273.96 | 0.996 183 | 10.872 6 | 5 260 | 400.71 | 0.996 595 |
| S-Y | 4 452.27 | 0.998 124 | 10.712 5 | 4 230 | 364.662 | 0.990 527 |
| BM | 3 900.91 | 0.884 219 | 7.023 68 | 3 830 | 180.009 | 0.995 676 |
| EG | 3 070.09 | 0.965 024 | 7.240 96 | 3 063 | 178.746 | 0.998 786 |
| TA | 912.77 | 0.978 809 | 7.514 6 | 910 | 72.96 | 0.999 898 |
| ZF | 5 244.04 | 0.957 637 | 8.139 92 | 5 195 | 210.571 | 0.995 141 |
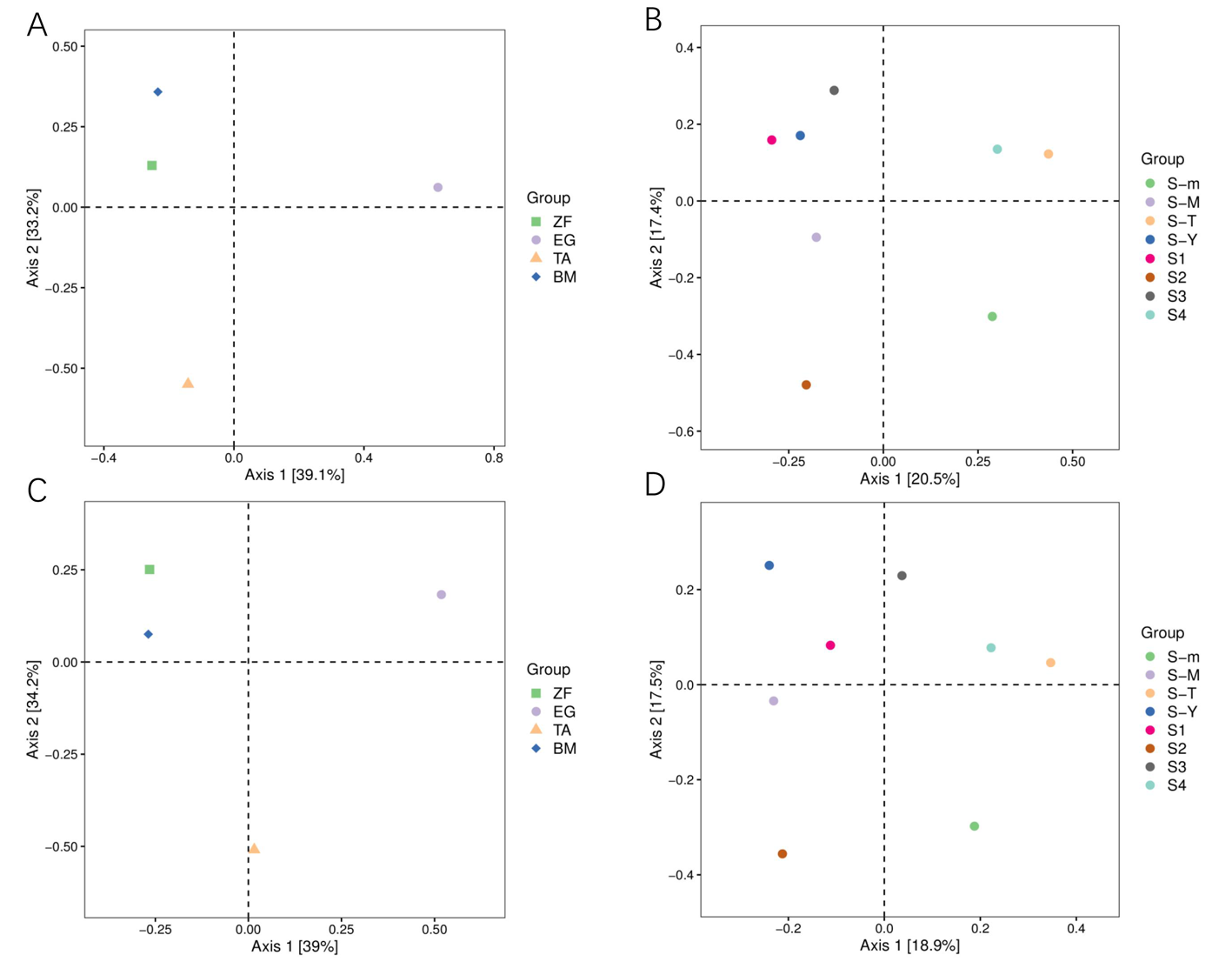
Fig.3 PCoA analysis of the upper reaches of the Tarim River Note:A:Analysis of PCoA based on Bray-Curtis about Water sample; B:Analysis of PCoA based on Bray-Curtis about Soil sample; C:Analysis of PCoA based on unweighted UniFrac about Water sample; D:Analysis of PCoA based on unweighted UniFrac about soil sample

Fig.8 Multiphase classification and identification of strain TRM49032 Note:A:system evolution relationship of strain TRM49032; B:morphological electron micrograph of strain TRM49032; C:cell wall polar lipid type of strain TRM49032(ninhydrin staining,anisaldehyde staining,molybdenum phosphate staining); D:cell wall fatty acid type table of strain TRM49032; E:Analysis of cell wall quinotype of strain TRM49032
| [1] | 刘海燕, 王敬敬, 赵维, 等. 塔里木河中下游流域棉田及胡杨林土壤细菌群落结构及多样性研究[J]. 微生物学通报, 2019, 46(9):2214-2230. |
| LIU Haiyan, WANG Jingjing, ZHAO Wei, et al. Structure of soil bacteria community and diversity in cotton field and Euphrates poplar forest in the middle and lower reaches of Tarim river basin[J]. Microbiology China, 2019, 46(9):2214-2230. | |
| [2] | 孙晨, 贡璐, 马勇刚, 等. 塔里木河上游典型绿洲土地利用时空格局分析[J]. 西北农林科技大学学报(自然科学版), 2020, 48(7):113-122. |
| SUN Chen, GONG Lu, MA Yonggang, et al. Temporal and spatial pattern analysis of land use in typical oasis in the upper reaches of the Tarim River[J]. Journal of Northwest A & F University(Natural Science Edition), 2020, 48(7):113-122. | |
| [3] | 罗建桦, 陶晔, 邢鹏. 湖泊微生物宏基因组学研究进展[J]. 湖泊科学, 2020, 32(1):271-280. |
|
LUO Jianhua, TAO Ye, XING Peng, et al. Mini-review:Advances of metagenomics research for lake microbiomes[J]. Journal of Lake Sciences, 2020, 32(1):271-280.
DOI URL |
|
| [4] | 吴庆龙, 邢鹏, 李化炳, 等. 草藻型稳态转换对湖泊微生物结构及其碳循环功能的影响[J]. 微生物学通报, 2013, 40(1):87-97. |
| WU Qinglong, XING Peng, LI Huabing, et al. Impacts of regime shift between phytoplankton and macrophyte on the microbial community structure and its carbon cycling in lakes[J]. Microbiology China, 2013, 40(1):87-97. | |
| [5] | Han D, Ha H K, Hwang C Y, et al. Bacterial communities along stratified water columns at the Chukchi Borderland in the western Arctic Ocean[J]. Deep-Sea Research Part II:Topical Studies in Oceanography, 2015, 120:52-60. |
| [6] | 吴庆龙, 江和龙. 中国湖泊微生物组研究[J]. 中国科学院院刊, 2017, 32(3):273-279. |
| WU Qinglong, JIANG Helong. China lake microbiome project[J]. Bulletin of Chinese Academy of Sciences, 2017, 32(3):273-279. | |
| [7] |
Ininbergs K, Bergman B, Larsson J, et al. Microbial metagenomics in the Baltic Sea:Recent advancements and prospects for environmental Monitoring[J]. Ambio, 2015, 44(Suppl 3):439-450.
DOI URL |
| [8] | 张庆芳, 杨超, 于爽, 等. 黄海海域海洋沉积物细菌多样性分析[J]. 微生物学通报, 2020, 47(2):370-378. |
| ZHANG Qingfang, YANG Chao, YU Shuang, et al. Bacterial diversity of marine sediments in the Yellow Sea[J]. Microbiology China, 2020, 47(2):370-378. | |
| [9] |
Vavourakis C D, Andrei A S, Mehrshad M, et al. A metagenomics roadmap to the uncultured genome diversity in hypersaline soda lake sediments[J]. Microbiome, 2018, 6(1):168.
DOI |
| [10] | 吴悦妮, 冯凯, 厉舒祯. 16S/18S/ITS扩增子高通量测序引物的生物信息学评估和改进[J]. 微生物学通报. 2020, 47(9):2897-2912. |
| WU Yueni, FENG Kai, LI Shuzhen, et al. In-silico evaluation and improvement on 16S/18S/ITS primers for amplicon high-throughput sequencing[J]. Microbiology China, 2020, 47(9):2897-2912. | |
| [11] |
Fung C, Rusling M, Lampeter T, et al. Automation of QIIME2 Metagenomic Analysis Platform[J]. Current Protocols. 2021. 1(9):e254.
DOI URL |
| [12] |
Ling L L, Schneider T, Peoples A J, et al. A new antibiotic kills pathogens without detectable resistance[J]. Nature, 2015, 517(7535):455-459.
DOI |
| [13] |
Sherpa R T, Reese C J, Montazeri A H. Application of iChip to Grow "Uncultivable" Microorganisms and its Impact on Antibiotic Discovery[J]. J Pharm Pharm Sci, 2015, 18(3):303.
PMID |
| [14] |
Lodhi A F, Zhang Y, Adil M, et al. Antibiotic discovery:combining isolation chip(iChip) technology and co-culture technique.[J]. Appl Microbiol Biotechnol, 2018, 102(17):7333-7341.
DOI |
| [15] | 阮继生, 黄英. 放线菌快速鉴定与系统分类[M]. 北京: 科学出版社, 2011. |
| RUAN Jisheng, HUANG Ying. Rapid identification and systematic classification of Actinomyces sp[M]. Beijing: Science Press, 2011. | |
| [16] | 雷艳娟. 骆驼蓬内生放线菌多样性及其次级代谢产物初探[D]. 阿拉尔: 塔里木大学, 2020. |
| LEI Yanjuan. The Diversity of Endophytic Actinobacteria from Peganum harmala and Primary Investigation of Their Secondary Metabolites[D]. Aral: Tarim University, 2020. | |
| [17] | 王扬. 一株糖霉菌新物种的多相分类及糖霉菌属CRISPR/Cas系统多样性分析[D]. 阿拉尔: 塔里木大学, 2018. |
| WANG Yang. The Polyphasic Taxonomy of a New Species of Glycomyces and Analyses of CRISPR/Cas System Diversity in Glycomyces[D]. Aral: Tarim University, 2018. | |
| [18] | 刘峰, 冯民权, 王毅博. 汾河入黄口夏季微生物群落结构分析[J]. 微生物学通报, 2019, 46(1):54-64. |
| LIU Feng, FENG Minquan, WANG Yibo. Microbial community structure of estuary of the Fenhe River into the Yellow River in summer[J]. Microbiology China, 2019, 46(1):54-64. | |
| [19] |
Kimura Z I, Okabe S. Acetate oxidation by syntrophic association between Geobacter sulfurreducens and a hydrogen-utilizing exoelectrogen[J] .ISME J, 2013, 7(8):1472-1482.
DOI URL |
| [20] |
Eyiuche N J, Asakawa S, Yamashita T, et al. Community analysis of biofilms on flame-oxidized stainless steel anodes in microbial fuel cells fed with different substrates[J]. BMC Microbiol, 2017, 17(1):145.
DOI PMID |
| [21] |
Ciok A, Budzik K, Zdanowski M K, et al. Plasmids of Psychrotolerant Polaromonas spp.Isolated From Arctic and Antarctic Glaciers - Diversity and Role in Adaptation to Polar Environments[J]. Front Microbiol, 2018, 9:1285.
DOI URL |
| [22] |
Gan H M, Lee Y P, Austin C M. Nanopore Long-Read Guided Complete Genome Assembly of Hydrogenophaga intermedia,and Genomic Insights into 4-Aminobenzenesulfonate,p-Aminobenzoic Acid and Hydrogen Metabolism in the Genus Hydrogenophaga[J]. Front Microbiol, 2017, 8:1880.
DOI URL |
| [23] |
Mu J, Cui X, Shao M J, et al. Microbial origin of bioflocculation components within a promising natural bioflocculant resource of Ruditapes philippinarum conglutination mud from an aquaculture farm in Zhoushan,China[J]. PLoS One, 2019, 14(6):e0217679.
DOI URL |
| [24] |
Vester J K, Glaring M A, Stougaard P. Improved cultivation and metagenomics as new tools for bioprospecting in cold environments[J]. Extremophiles, 2015, 19(1):17-29.
DOI PMID |
| [25] |
Yang Q, Cai S, Dong S W, et al. Biodegradation of 3-methyldiphenylether(MDE) by Hydrogenophaga atypical strain QY7-2 and cloning of the methy-oxidation gene mdeABCD[J]. Sci Rep., 2016, 6:39270.
DOI |
| [26] |
Zhao T, Hu K, Li J, et al. Current insights into the microbial degradation for pyrethroids:strain safety,biochemical pathway,and genetic engineering[J]. Chemosphere, 2021, 279:130542.
DOI URL |
| [27] |
Zhou S L, Sun Y, Yu M H, et al. Linking Shifts in Bacterial Community Composition and Function with Changes in the Dissolved Organic Matter Pool in Ice-Covered Baiyangdian Lake,Northern China[J]. Microorganisms., 2020, 8(6):883.
DOI URL |
| [28] | 肖方南, 姜梦, 李媛媛, 等. 塔里木河下游柽柳灌丛土壤真菌群落结构及多样性分析[J]. 干旱区地理, 2021, 44(3):759-768. |
| XIAO Fangnan, JIANG Meng, LI Yuanyuan, et al. Community structure and diversity of soil fungi in Tamarix chinensis shrubs in the lower reaches of Tarim River[J]. Arid Land Geography, 2021, 44(3):759-768. | |
| [29] | 李媛媛. 塔里木河下游胡杨根际微生物群落结构与土壤理化性质的相关性分析[D]. 石河子: 石河子大学, 2022. |
| LI Yuanyuan. Structure of Soil Microbial Community in the Rhizosphere of Populus euphratica in the Lower Reaches of Tarim River and Its Correlation with Soil Physicochemical Properties[D]. Shihezi: Shihezi University, 2022. |
| [1] | ZENG Wanying, GENG Hongwei, CHENG Yukun, LI Sizhong, QIAN Songting, GAO Weishi, ZHANG Liming. Comprehensive evaluation of drought resistance during the rapid growth stage of sugar beet cultivars [J]. Xinjiang Agricultural Sciences, 2024, 61(9): 2140-2151. |
| [2] | XU Maomao, GAO Jie, LI Junming, LI Xin, LIU Lei, PAN Feng. Population diversity analysis of 20 commercial tomato cultivars [J]. Xinjiang Agricultural Sciences, 2024, 61(9): 2191-2196. |
| [3] | MIAO Yu, CHEN Cuixia, MA Yanming, XING Guofang, DONG Yusheng, CHEN Zhijun, WANG Xian, XIANG Li. Genetic diversity analysis of phenotypic traits of 276 Central Asian barley germplasm resources [J]. Xinjiang Agricultural Sciences, 2024, 61(8): 1888-1895. |
| [4] | WANG Qingpeng, YAN Chengcai, WANG Zhe, GOU Changqing, WANG Lan, FENG Hongzu, HAO Haiting. Anther bacterial diversity of Korla Fragrant Pear before and after flowering [J]. Xinjiang Agricultural Sciences, 2024, 61(8): 1976-1982. |
| [5] | LIU Huifang, HAN Hongwei, ZHUANG Hongmei, WANG Qiang, GAO Axiang, WANG Hao. Effects of increased application of microbial fertilizer on soil microbial diversity of cucumber in facilities [J]. Xinjiang Agricultural Sciences, 2024, 61(7): 1727-1737. |
| [6] | YANG Lu, WANG Na, FAN Shaoli, CHENG Ping, LI Hong, WANG Yangdong. Analysis of phenotypic trait variation characteristics of Morus nigra L.germplasm resources [J]. Xinjiang Agricultural Sciences, 2024, 61(5): 1172-1181. |
| [7] | GENG Meiju, WANG Xinhui, LIU Xiaoying, LYU Pei. Effects of sealing on fungal communities in desert grasslands [J]. Xinjiang Agricultural Sciences, 2024, 61(5): 1250-1258. |
| [8] | Reyihan Abulizi, HE Xuemin, YANG Huan, HUANG Pengcheng, FENG Haipeng, WANG Yongzhi. Characteristics of chlorophyll-fluorescence parameters of Suaeda microphylla Pall.and their responses to soil factors in different water-salt habitats [J]. Xinjiang Agricultural Sciences, 2024, 61(2): 485-494. |
| [9] | WANG Fan, LI Yushan, WANG Wei, DENG Chaohong, ZHAO Lianjia, MA Yue, XIAO Jing, ZHUANG Hongmei, Xu Hongjun. Genetic diversity analysis of major nutritional growth traits in 74 turnip germplasm resources [J]. Xinjiang Agricultural Sciences, 2024, 61(11): 2601-2613. |
| [10] | LI Chao, YANG Ying, ZHENG Heyun, YANG Jianli, CHEN Wei, YANG Mi, SUN Yuping. Genetic diversity and population structure analysis of melon germplasm resources in Xinjiang based on SSR fluorescence markers [J]. Xinjiang Agricultural Sciences, 2024, 61(11): 2614-2625. |
| [11] | HUANG Xinglei, WANG Weiran, WANG Meng, ZHU Jiahui, LIN Feng, QIN Guoli, YANG Jing, Alifu Aierxi, WU Quanzhong, KONG Jie. Diversity evaluation of machine-picked agronomic traits in Gossypium barbadense L. germplasm resources [J]. Xinjiang Agricultural Sciences, 2024, 61(1): 1-8. |
| [12] | XI Ouyan, WANG Chenri, Gulinuer Tulaxi, HU Hongying. Characteristics of pollinating insect types and floral visiting behavior in wild fruit forests of the Ili Valley [J]. Xinjiang Agricultural Sciences, 2024, 61(1): 190-198. |
| [13] | Wang Tianling, Hou Xianfei, Shi Junjie, Sun Quanxi, Jia Donghai, Gu Yuanguo, Shan Shihua, Miao Haocui, Li Qiang. Genetic diversity analysis of 67 creeping peanut germplasm resources [J]. Xinjiang Agricultural Sciences, 2024, 61(1): 42-54. |
| [14] | WANG Dandan, LI Yan, ZHANG Qingyin, Li Shidong, PANG Yongchao, MA Kunzhi, MA Long, NIU Ruisheng, ZHONG Zengming, QI Lianfen, SHI Jianhua. Effects of different microbial treatments on tomato soil microbial diversity [J]. Xinjiang Agricultural Sciences, 2023, 60(9): 2248-2257. |
| [15] | ZHOU Shijie, DONG Yiqiang, Asitaiken Julihaiti, NIE Tingting, JIANG Anjing, AN Shazhou. Quantitative characteristics and diversity of sagebrush desert plant communities on the northern slope of Tianshan Mountains [J]. Xinjiang Agricultural Sciences, 2023, 60(9): 2298-2305. |
| Viewed | ||||||
|
Full text |
|
|||||
|
Abstract |
|
|||||