

Xinjiang Agricultural Sciences ›› 2022, Vol. 59 ›› Issue (6): 1312-1330.DOI: 10.6048/j.issn.1001-4330.2022.06.002
• Crop Genetics and Breeding·Cultivation Physiology·Physiology and Biochemistry • Previous Articles Next Articles
SONG Jikun1,2( ), XIN Yue1, LI Longyun1, LIU Guoyuan1, PEI Wenfeng1,2, MA Jianjiang1, QU Yanying2, YU Jiwen1,2(
), XIN Yue1, LI Longyun1, LIU Guoyuan1, PEI Wenfeng1,2, MA Jianjiang1, QU Yanying2, YU Jiwen1,2( ), WU Man1(
), WU Man1( )
)
Received:2021-09-26
Online:2022-06-20
Published:2022-07-07
Correspondence author:
YU Jiwen, WU Man
Supported by:
宋吉坤1,2( ), 辛玥1, 李龙云1, 刘国元1, 裴文锋1,2, 马建江1, 曲延英2, 于霁雯1,2(
), 辛玥1, 李龙云1, 刘国元1, 裴文锋1,2, 马建江1, 曲延英2, 于霁雯1,2( ), 吴嫚1(
), 吴嫚1( )
)
通讯作者:
于霁雯,吴嫚
作者简介:宋吉坤(1995-),男,山东青岛人,博士研究生,研究方向为棉花分子育种,(E-mail) sjkow513@163.com
基金资助:CLC Number:
SONG Jikun, XIN Yue, LI Longyun, LIU Guoyuan, PEI Wenfeng, MA Jianjiang, QU Yanying, YU Jiwen, WU Man. Comparative Analysis of Gene Expression between Giza 75 and SG 747 Using Cotton Oligonucleotide Microarrays during Fiber Development[J]. Xinjiang Agricultural Sciences, 2022, 59(6): 1312-1330.
宋吉坤, 辛玥, 李龙云, 刘国元, 裴文锋, 马建江, 曲延英, 于霁雯, 吴嫚. 利用基因芯片分析比较Giza75和SG747纤维发育差异表达基因[J]. 新疆农业科学, 2022, 59(6): 1312-1330.
Add to citation manager EndNote|Ris|BibTeX
URL: https://www.xjnykx.com/EN/10.6048/j.issn.1001-4330.2022.06.002
| 探针编号 Probe Set | NCBI 登录号 Accessionnumber | 基因名称 Gene Name | 引物序列 Sequence | 引物长度 Length (bp) |
|---|---|---|---|---|
| __ | AY305737 | Actin9 | -F5’-ATCCTCCGTCTTGACCTTG -3’ | 19 |
| -R5’-TGTCCGTCAGGCAACTCAT-3’ | 19 | |||
| Ghi.8448.1.S1_x_at | AF521240.1 | GhXu-142 βTUB1 | -F5’-TAACTATGTCGGCACTTC -3’ | 18 |
| -R5’-AATCAATTCAGCTCCTTC -3’ | 18 | |||
| Gra.2198.1.A1_at | CO100609 | GhSAHH | -F5’-CAGCTCCCAGATCCGTCTTC -3’ | 20 |
| -R5’-CACCAACTAATCTCTCCCTCATCC -3’ | 24 | |||
| Gra.85.1.S1_at | CO097359 | GhCESA4 | -F5’- GTAACCATAAGGCATGTCCAAAATG-3’ | 25 |
| -R5’- AGAAAAGGTGGGAATGAACGAA-3’ | 22 | |||
| Ghi.249.1.A1_at | DQ204496.1 | GhAlpha-expansin 2 | -F5’-CCCCTGTATGAAGAAAGGAGGA -3’ | 22 |
| -R5’-AACATCTCCAGCACCACCAAC -3’ | 21 | |||
| -R5’-CGAAGTTGTTGGCAGCGTCT -3’ | 20 |
Table 1 Gene Primers for quantitative RT-PCR analysis
| 探针编号 Probe Set | NCBI 登录号 Accessionnumber | 基因名称 Gene Name | 引物序列 Sequence | 引物长度 Length (bp) |
|---|---|---|---|---|
| __ | AY305737 | Actin9 | -F5’-ATCCTCCGTCTTGACCTTG -3’ | 19 |
| -R5’-TGTCCGTCAGGCAACTCAT-3’ | 19 | |||
| Ghi.8448.1.S1_x_at | AF521240.1 | GhXu-142 βTUB1 | -F5’-TAACTATGTCGGCACTTC -3’ | 18 |
| -R5’-AATCAATTCAGCTCCTTC -3’ | 18 | |||
| Gra.2198.1.A1_at | CO100609 | GhSAHH | -F5’-CAGCTCCCAGATCCGTCTTC -3’ | 20 |
| -R5’-CACCAACTAATCTCTCCCTCATCC -3’ | 24 | |||
| Gra.85.1.S1_at | CO097359 | GhCESA4 | -F5’- GTAACCATAAGGCATGTCCAAAATG-3’ | 25 |
| -R5’- AGAAAAGGTGGGAATGAACGAA-3’ | 22 | |||
| Ghi.249.1.A1_at | DQ204496.1 | GhAlpha-expansin 2 | -F5’-CCCCTGTATGAAGAAAGGAGGA -3’ | 22 |
| -R5’-AACATCTCCAGCACCACCAAC -3’ | 21 | |||
| -R5’-CGAAGTTGTTGGCAGCGTCT -3’ | 20 |
| 性状 Trait | 基因型Genetype | T测验T test | |||
|---|---|---|---|---|---|
| Giza75 | SG7474 | T Stat | P(T≤T)双尾 P(T≤t) two- tailed | ||
| 纤维长度 Fiber Length (mm) | 34.090 0 | 29.201 7 | 10.433 0 | 0.000 0 | ** |
| 整齐度指数 Uniformity index(%) | 86.066 7 | 84.666 7 | 2.613 8 | 0.028 1 | * |
| 马克隆值 Micronair value | 4.565 0 | 5.621 7 | -3.038 0 | 0.028 8 | * |
| 伸长率 Fiber Elongation rate(%) | 5.750 0 | 6.416 7 | -1.913 0 | 0.092 1 | |
| 纤维强度 Fiber Strength (cN/tex) | 40.391 3 | 27.693 3 | 8.487 3 | 0.000 0 | ** |
| 籽棉产量 Seed Yield (kg/667m2) | 70.947 5 | 191.813 8 | -4.838 0 | 0.000 7 | ** |
| 皮棉产量 Lint Yield (kg/667m2) | 26.677 5 | 78.272 5 | -5.397 7 | 0.000 2 | ** |
| 衣分 Lint Percentage (%) | 36.926 3 | 41.707 5 | -4.312 8 | 0.000 8 | ** |
| 铃重 Boll Weight (g) | 3.617 5 | 6.007 5 | -10.792 4 | 0.000 0 | ** |
Table 2 Fiber quality andyield difference between G. hirsutum and G. barbadense
| 性状 Trait | 基因型Genetype | T测验T test | |||
|---|---|---|---|---|---|
| Giza75 | SG7474 | T Stat | P(T≤T)双尾 P(T≤t) two- tailed | ||
| 纤维长度 Fiber Length (mm) | 34.090 0 | 29.201 7 | 10.433 0 | 0.000 0 | ** |
| 整齐度指数 Uniformity index(%) | 86.066 7 | 84.666 7 | 2.613 8 | 0.028 1 | * |
| 马克隆值 Micronair value | 4.565 0 | 5.621 7 | -3.038 0 | 0.028 8 | * |
| 伸长率 Fiber Elongation rate(%) | 5.750 0 | 6.416 7 | -1.913 0 | 0.092 1 | |
| 纤维强度 Fiber Strength (cN/tex) | 40.391 3 | 27.693 3 | 8.487 3 | 0.000 0 | ** |
| 籽棉产量 Seed Yield (kg/667m2) | 70.947 5 | 191.813 8 | -4.838 0 | 0.000 7 | ** |
| 皮棉产量 Lint Yield (kg/667m2) | 26.677 5 | 78.272 5 | -5.397 7 | 0.000 2 | ** |
| 衣分 Lint Percentage (%) | 36.926 3 | 41.707 5 | -4.312 8 | 0.000 8 | ** |
| 铃重 Boll Weight (g) | 3.617 5 | 6.007 5 | -10.792 4 | 0.000 0 | ** |
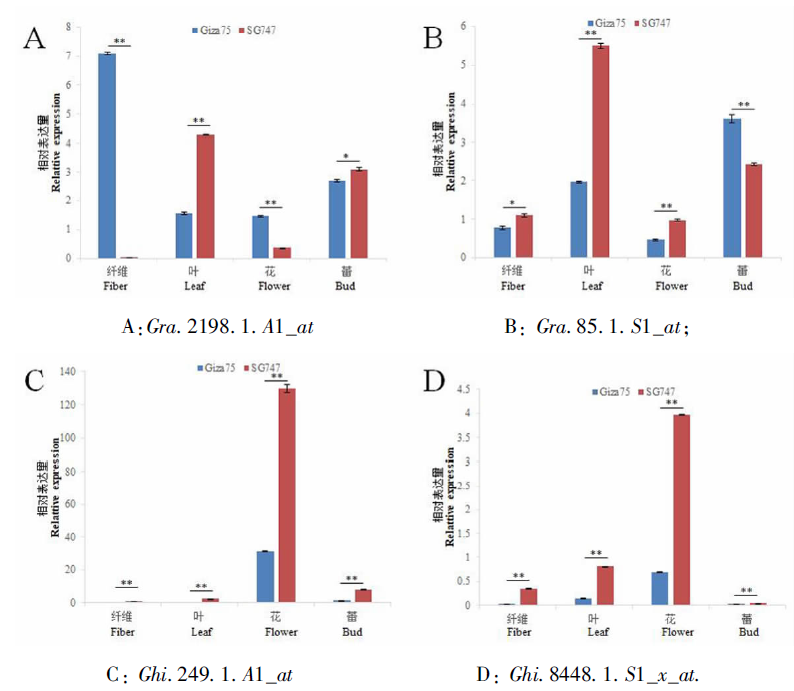
Fig.3 qRT-PCR analysis results of differentially expressed genes in different tissues. Note: The fiber is the fiber at 5 days after flowering; * and ** indicate significant differences at 0.05 and 0.01 levels, respectively.
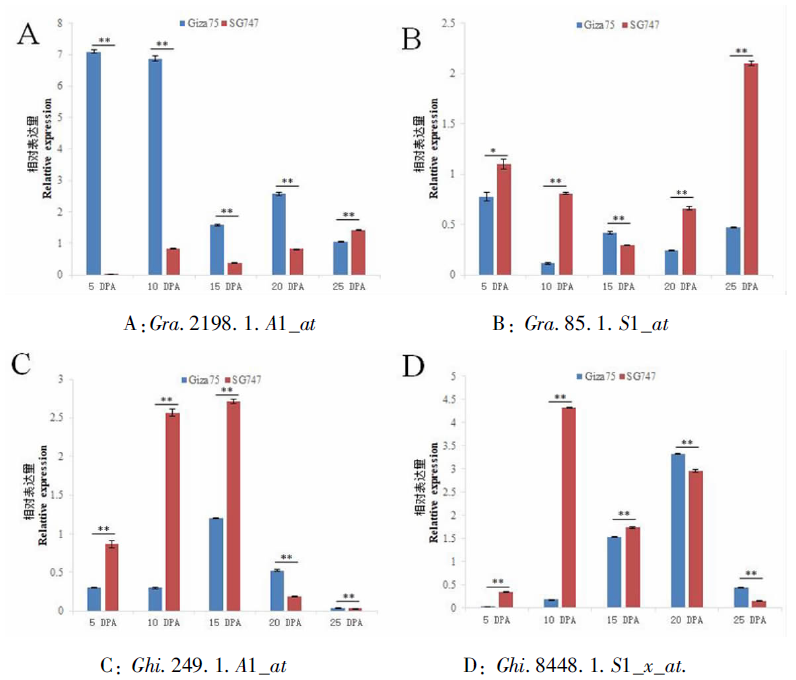
Fig.4 qRT-PCR analysis results of differentially expressed genes at different developmental stages of fiber Note: DPA is the number of days after flowering; * and ** indicate significant differences at 0.05 and 0.01 levels, respectively
| [1] | Wendel J F, Brubaker C, Alvarez I, et al. Evolution and natural history of the cotton genus[J]. In: Paterson AP (ed.) Genetics and Genomics of Cotton, 2009, 3-22. |
| [2] | Bowman D T, Gutierrez O A. Sources of fiber strength in the U.S. upland cotton crop from 1980 to 2000[J]. The Journal of Cotton Science, 2003, 7:164-169. |
| [3] |
Sajid M, Ahmad R I, Muhammad A R, et al. Role of SNPs in determining QTLs for major traits in cotton[J]. Journal of Cotton Research, 2019, 2:5.
DOI URL |
| [4] |
Ma L L, Su Y, Nie H S, et al. QTL and genetic analysis controlling fiber quality traits using paternal back cross population in upland cotton[J]. Journal of Cotton Research, 2020, 3:22.
DOI URL |
| [5] |
Deng X Y, Gong J W, Liu A Y, et al. QTL mapping for fiber quality and yield related traits across multiple generations in segregating population of CCRI 70[J]. Journal of Cotton Research, 2019, 2:13.
DOI URL |
| [6] |
Tu L L, Zhang X L, Liang S G, et al. Genes expression analyses of sea-island cotton (Gossypium barbadense L.) during fiber development[J]. Plant Cell Reports, 2007, 26(8):1309-1320.
PMID |
| [7] |
Wu M, Zhang L Y, Li X L, et al. A comparative transciptome analysis of two sets of backcross inbred lines differing in lint-yield derived from a Gossypium hirsutum × Gossypium barbadense population[J]. Molecular Genetics and Genomics, 2016, 291(4):1749-1767.
DOI URL |
| [8] |
Alabady M S, Youn E, Wilkins T A. Double feature selection and cluster analyses in mining of microarray data from cotton[J]. BMC Genomics, 2008, 9:295.
DOI PMID |
| [9] |
Wu M, Li L Y, Liu G Y, et al. Differentially expressed genes between two groups of backcross inbred lines differing in fiber length developed from Upland × Pima cotton[J]. Molecular Biology Reports, 2019, 46(1):1199-1212.
DOI URL |
| [10] |
Qin Y M, Zhu Y X. How cotton fibers elongate: a tale of linear cell-growth mode[J]. Current Opinion in Plant Biology, 2011, 14(1):106-11.
DOI URL |
| [11] |
Liu C X, Li Z F, Dou L L, et al. A genome-wide identification of the BLH gene family reveals BLH1 involved in cotton fiber development[J]. Journal of Cotton Research, 2020, 3:26.
DOI URL |
| [12] |
Chen F, Guo Y J, Chen L, et al. Global identification of genes associated with xylan biosynthesis in cotton fiber[J]. Journal of Cotton Research, 2020, 3:25.
DOI URL |
| [13] |
Yu J W, Zhang K, Li S Y, et al. Mapping quantitative trait loci for lint yield and fiber quality across environments in a Gossypium hirsutum × Gossypium barbadense backcross inbred line population[J]. Theoretical and Applied Genetics, 2013, 126(1): 275-287.
DOI URL |
| [14] | 蒋建雄, 张天真. 利用CTAB/酸酚法提取棉花组织总RNA[J]. 棉花学报, 2003, 15(3):166-167. |
| JIANG Jiangxiong, ZHANG Tianzhen. Extraction of total RNA in cotton tissues with CTAB-acidic phenolic method[J]. Cotton Science, 2003, 15(3):166-167 | |
| [15] | 李龙云, 于霁雯, 翟红红, 等. 利用基因芯片技术筛选棉纤维伸长相关基因[J]. 作物学报, 2011, 37(1):95-104. |
|
LI Longyun, YU Jiwen, ZHAI Honghong, et al. Identification of fiber length related genes using cotton oligonucleotide microarrays[J]. Acta Agronomica Sinica, 2011, 37(1): 95-104.
DOI URL |
|
| [16] | 吴嫚, 李龙云, 裴文锋, 等. 利用马克隆值差异显著的BILs鉴定棉纤维品质相关基因[J]. 棉花学报, 2020, 32(1):52-62. |
| WU Man, LI Longyun, PEI Wenfeng, et al. Identification of differentially expressed genes in developing cotton fibers between two groups of backcross inbred lines differing in fiber Micronaire[J]. Cotton Science, 2020, 32(1):52-62. | |
| [17] |
Hu Y, Chen J D, Fang L, et al. Gossypium barbadense and Gossypium hirsutum genomes provide insights into the origin and evolution of allotetraploid cotton[J]. Nature Genetics, 2019, 51(4):739-748.
DOI URL |
| [18] |
Chee P, Draye X, Jiang C X, et al. Molecular dissection of interspecific variation between Gossypium hirsutum and Gossypium barbadense (cotton) by a backcross-self approach: I. fiber elongation[J]. Theoretical and Applied Genetics, 2005, 111(4):757-763.
DOI URL |
| [19] | 李龙云. 利用基因芯片技术筛选棉纤维发育相关基因[D]. 杨凌: 西北农林科技大学, 2010. |
| LI Longyun. Identification Of Fiber Quality Genes By Using Cotton Oligonucleotide Microarrays with Gene Expression Analysis[D]. Yangling: Northwest A&F University, 2010. | |
| [20] |
Sun Q, Cai Y F, Xie Y F, et al. Gene expression profiling during gland morphogenesis of a mutant and a glandless upland cotton[J]. Molecular Biology Reports, 2010, 37(7):3319-3325.
DOI URL |
| [21] | 佘义斌, 朱一超, 张天真, 等. 棉花腺苷高半胱氨酸水解酶cDNA的克隆、表达及染色体定位[J]. 作物学报, 2008, (6):958-964. |
|
SHE Yibin, ZHU Yichao, ZHANG Tianzhen, et al. Cloning, expression, and mapping of s-adenosyl-l-homocysteine hydrolase (GhSAHH) cDNA in cotton[J]. Acta Agronomica Sinica, 2008, 34(6):958-964.
DOI URL |
|
| [22] |
Li C H, Zhu Y Q, Meng Y L, et al. Isolation of genes preferentially expressed in cotton fibers by cDNA filter arrays and RT-PCR[J]. Plant Science, 2002, 163(6):1113-1120.
DOI URL |
| [23] |
Bach I. The LIM domain: regulation by association[J]. Mechanisms of Development, 2000, 91(1-2):5-17.
PMID |
| [24] |
Saxena I M, Brown R M. Cellulose synthases and related enzymes[J]. Current Opinion in Plant Biology, 2000, 3(6):523-31.
PMID |
| [25] | 武耀廷, 张恒木, 刘进元. 棉纤维细胞发育过程中纤维素的生物合成[J]. 棉花学报, 2003, 15(3):174-179. |
| WU Yaoting, ZHANG Hengmu, LIU Jinyuan. Cellulose biosynthesis in developing cotton fibers[J]. Cotton Science, 2003, 15(3):174-179. | |
| [26] |
Zhang S J, Jiang Z X, Chen J, et al. The cellulose synthase (CesA) gene family in four Gossypium species: phylogenetics, sequence variation and gene expression in relation to fiber quality in Upland cotton[J]. Molecular Genetics and Genomics, 2021, 296(2):355-368.
DOI URL |
| [27] |
Harmer S E, Oxford S J, Timmis J N. Characterization of six alpha-expansin genes in Gossypium hirsutum (upland cotton)[J]. Molecular Genetics and Genomics, 2002, 268(1):1-9.
PMID |
| [28] | Wilkins T A, Arpat A, Sickler B. Cotton fiber genomics: developmental mechanisms[J]. Pflanzenschutz-Nachrichten Bayer, 2005, 1:119-139. |
| [29] |
Gou J Y, Wang L J, Chen S P, et al. Gene expression and metabolite profiles of cotton fiber during cell elongation and secondary cell wall synthesis[J]. Cell Research, 2007, 17(5):422-34.
DOI URL |
| [30] |
Li Y, Tu L L, Pettolino F A, et al. GbEXPATR, a species-specific expansin, enhances cotton fibre elongation through cell wall restructuring[J]. Plant Biotechnology Journal, 2016, 14(3):951-963.
DOI URL |
| [31] | 杨君, 马峙英, 王省芬. 棉花纤维品质改良相关基因研究进展[J]. 中国农业科学, 2016, 49(22):4310-4322,4469-4472. |
| YANG Jun, MA Zhiying, WANG Xingfen. Progress in studies on genes related to fiber quality improvement of cotton[J]. Scientia Agricultura Sinica, 2016, 49(22):4310-4322,4469-4472. | |
| [32] |
Dixon D C, Seagull R W, Triplett B A. Changes in the accumulation of α- and β-tubulin isotypes during cotton fiber development[J]. Plant Physiology, 1994, 105(4):1347-1353.
PMID |
| [33] |
Li X B, Cai L, Cheng N H, et al. Molecular characterization of the cotton GhTUB1 gene that is preferentially expressed in fiber[J]. Plant Physiology, 2002, 130(2), 666-674.
DOI URL |
| [34] | 刘迪秋. 陆地棉纤维特异表达基因的克隆与表达研究[D]. 武汉: 华中农业大学, 2007. |
| LIU Diqiu. Molecular cloning and expression analysis of the genes specifically expressed in fibers of Gossypium hirsutum[D]. Wuhan: Huazhong Agricultural University, 2007 |
| [1] | ZHANG Tingjun, LI Zihui, CUI Yujiang, SUN Xiaogui, CHEN Fang. Effects of microbial agents on cotton growth and soil physico-chemical properties [J]. Xinjiang Agricultural Sciences, 2024, 61(9): 2269-2276. |
| [2] | ZHANG Chengjie, HU Haoran, DUAN Songjiang, WU Yifan, ZHANG Jusong. Effects of nitrogen-dense interaction on growth, development, yield and quality of Gossypium barbadense L. [J]. Xinjiang Agricultural Sciences, 2024, 61(8): 1821-1830. |
| [3] | DONG Zhenlin, WAN Sumei, XIONG Shiwu, MA Yunzhen, MAO Tingyong, YANG Beifang, LUO Lei, LIU Chaoqun, CHEN Guodong, LI Yabing. Effects of different planting densities on agronomic traits and yield of Zhongmian 113 [J]. Xinjiang Agricultural Sciences, 2024, 61(5): 1102-1111. |
| [4] | LIU Taijie, CHEN Bing, YANG Li, WANG Jing, ZHAO Jing, LI Xiang, TANG Guanglan, WANG Gang, HAN Huanyong, WANG Fangyong. Effects of different spraying machinery and pesticide combination on the defoliation,the ripeness,yield and quality of cotton [J]. Xinjiang Agricultural Sciences, 2024, 61(4): 852-860. |
| [5] | HE Wanjie, MENG Hanying, ZHI Mengting, CHEN Jing. Analysis of the antennal transcriptome and differentially expressed genes of female and male Monolepta signata [J]. Xinjiang Agricultural Sciences, 2024, 61(4): 984-995. |
| [6] | WANG Kaidi, GAO Chenxu, PEI Wenfeng, YANG Shuxian, ZHANG Wenqing, SONG Jikun, MA Jianjiang, WANG Li, YU Jiwen, CHEN Quanjia. Identification of TRM gene family and fiber quality related excellent haplotype analysis in Gossypium hirsutum L. [J]. Xinjiang Agricultural Sciences, 2024, 61(3): 521-536. |
| [7] | WANG Wei, ZHANG Renfu, LIU Haiyang, DING Ruifeng, LIANG Gemei, YAO Ju. Transcriptome analysis of Aphis gossypii sulfoxaflor and acetamiprid-resistant strains with different genetic backgrounds of resistance [J]. Xinjiang Agricultural Sciences, 2024, 61(12): 3078-3088. |
| [8] | HUANG Xinglei, WANG Weiran, WANG Meng, ZHU Jiahui, LIN Feng, QIN Guoli, YANG Jing, Alifu Aierxi, WU Quanzhong, KONG Jie. Diversity evaluation of machine-picked agronomic traits in Gossypium barbadense L. germplasm resources [J]. Xinjiang Agricultural Sciences, 2024, 61(1): 1-8. |
| [9] | WANG Xin, LIN Tao, CUI Jianping, WU Fengquan, TANG Zhixuan, CUI Laiyuan, GUO Rensong, WANG Liang, ZHENG Zipiao. Effects of planting mode and irrigation quota on yield and fiber quality of machine-picked long-staple cotton [J]. Xinjiang Agricultural Sciences, 2023, 60(8): 1821-1829. |
| [10] | DUAN Songjiang, PENG Zengying, SHEN Yingying, Mulidier Baibolati, WU Yifan, CUI jianping, ZHANG Jusong. Responses of seed cotton yield and fiber quality of different sea island cotton varieties to nitrogen fertilizer [J]. Xinjiang Agricultural Sciences, 2023, 60(7): 1569-1579. |
| [11] | WANG Wentao, WU Bo, TAI Hongzhong, LIAN Wenming, DAI Cuirong, LI Shuangjiang, PU Yanmei. Effects of different sowing dates on cotton growth in aral reclamation area, Xinjiang [J]. Xinjiang Agricultural Sciences, 2023, 60(6): 1413-1422. |
| [12] | LIU Chenxi, ZHU Yuting, ZHOU Qiang, CHEN Jin, ZHAO Wenjie, ZHENG Kai. Cloning and bioinformatics analysis of β-carotene isomerase GbD27-6 gene in sea island cotton [J]. Xinjiang Agricultural Sciences, 2023, 60(12): 2869-2877. |
| [13] | MA Jun, YANG Yanlong, SHI Weijun, WANG Penglong, ZHENG Juyun, GUO Rensong, HU Wenran, YANG Yang. Analysis of QTL Mapping for Fiber Quality Traits in Introgression Lines Cotton [J]. Xinjiang Agricultural Sciences, 2023, 60(1): 11-16. |
| [14] | Kadliya Abudukelimu, ZHANG Jusong, CHEN Zhen, HE Hongwei, CUI Jianping, LIN Tao, GUO Rensong. Effects of Plant Spacing and Nitrogen Fertilizer Management on the Growth and Yield of Machine-Picked Cotton [J]. Xinjiang Agricultural Sciences, 2022, 59(2): 269-278. |
| [15] | GUO Xiaoyan, SUN Guilan, XIONG Shiwu, CHEN Huanxuan, HAN Yingchun, WANG Guoping, LI Cundong, LI Yabing, ZHANG Yongjiang, WANG Zhanbiao. Effects of Nitrogen Application Rates on Nutrition Uptake and Utilization, Yield and Fiber Quality of Cotton [J]. Xinjiang Agricultural Sciences, 2021, 58(7): 1246-1254. |
| Viewed | ||||||
|
Full text |
|
|||||
|
Abstract |
|
|||||