

Xinjiang Agricultural Sciences ›› 2024, Vol. 61 ›› Issue (4): 984-995.DOI: 10.6048/j.issn.1001-4330.2024.04.023
• Plant Protection·Microbes·Agricultural Equipment Engineering and Mechanization • Previous Articles Next Articles
HE Wanjie( ), MENG Hanying, ZHI Mengting, CHEN Jing(
), MENG Hanying, ZHI Mengting, CHEN Jing( )
)
Received:2023-08-19
Online:2024-04-20
Published:2024-05-31
Correspondence author:
CHEN Jing
Supported by:通讯作者:
陈静
作者简介:何婉洁(1997-),女,新疆人,硕士研究生,研究方向为农业昆虫与害虫防治,(E-mail)1287336486@qq.com
基金资助:CLC Number:
HE Wanjie, MENG Hanying, ZHI Mengting, CHEN Jing. Analysis of the antennal transcriptome and differentially expressed genes of female and male Monolepta signata[J]. Xinjiang Agricultural Sciences, 2024, 61(4): 984-995.
何婉洁, 孟涵颖, 支梦婷, 陈静. 双斑长跗萤叶甲雌虫、雄虫触角转录组及差异表达基因分析[J]. 新疆农业科学, 2024, 61(4): 984-995.
| 基因ID Gene ID | 基因名称 Gene name | 上游引物(5’-3’) Forward | 下游引物(3’-5’) Reverse | 扩增片段长度 Product length |
|---|---|---|---|---|
| Cluster-12689.30761 | MsigOBP2 | TGGTTTCAGCGAACGCAATC | TCCTTCATGACTGTCTGGTTCC | 152 |
| Cluster-12689.31568 | MsigOBP3 | TAGAGGCAAATGCACCACCAT | TAGAGGCAAATGCACCACCAT | 194 |
| Cluster-12689.26420 | MsigOBP5 | AAATGCGGATGGCTCCATGA | TCCCAGTGCTCGTCCTTCTA | 199 |
| Cluster-12689.31318 | MsigOBP6 | GCAACCCATTTGTTGCCTCA | TGTACGTGTGTCGGACAGTG | 165 |
| Cluster-12689.8427 | MsigOBP8 | CATCCATGCGCCTTTCAAGA | GCCAAAGAAGAGTGTCAAGCTG | 199 |
| Cluster-12689.26435 | MsigOBP15 | TGGGAATCTTAGCTGGTGGC | CACACCAATTTCATCGAAAACGC | 191 |
| Cluster-12689.34706 | MsigOBP18 | AGGACTCCAAAATGAAGACGGT | ACAAGGACACGTTTGGGGTT | 189 |
| Cluster-12689.31354 | MsigOBP20 | ACGGTTGCGTCGTCTATTCC | AAGATCCAGAAACACGAGCGG | 181 |
| Cluster-12689.30900 | MsigOBP21 | TGGACACCAAGGCATCTCAC | TCTGAGGCGCCTGAATAACA | 196 |
Tab. 1 The information about primer of olfactory differently expressed genes
| 基因ID Gene ID | 基因名称 Gene name | 上游引物(5’-3’) Forward | 下游引物(3’-5’) Reverse | 扩增片段长度 Product length |
|---|---|---|---|---|
| Cluster-12689.30761 | MsigOBP2 | TGGTTTCAGCGAACGCAATC | TCCTTCATGACTGTCTGGTTCC | 152 |
| Cluster-12689.31568 | MsigOBP3 | TAGAGGCAAATGCACCACCAT | TAGAGGCAAATGCACCACCAT | 194 |
| Cluster-12689.26420 | MsigOBP5 | AAATGCGGATGGCTCCATGA | TCCCAGTGCTCGTCCTTCTA | 199 |
| Cluster-12689.31318 | MsigOBP6 | GCAACCCATTTGTTGCCTCA | TGTACGTGTGTCGGACAGTG | 165 |
| Cluster-12689.8427 | MsigOBP8 | CATCCATGCGCCTTTCAAGA | GCCAAAGAAGAGTGTCAAGCTG | 199 |
| Cluster-12689.26435 | MsigOBP15 | TGGGAATCTTAGCTGGTGGC | CACACCAATTTCATCGAAAACGC | 191 |
| Cluster-12689.34706 | MsigOBP18 | AGGACTCCAAAATGAAGACGGT | ACAAGGACACGTTTGGGGTT | 189 |
| Cluster-12689.31354 | MsigOBP20 | ACGGTTGCGTCGTCTATTCC | AAGATCCAGAAACACGAGCGG | 181 |
| Cluster-12689.30900 | MsigOBP21 | TGGACACCAAGGCATCTCAC | TCTGAGGCGCCTGAATAACA | 196 |
| 样品 Sample | 原始读数 Raw reads | 筛选后读数 Clean reads | 总核酸数(G) Clean bases | 错误率 Error rate (%) | Q20 (%) | Q30 (%) | GC比值 GC content (%) |
|---|---|---|---|---|---|---|---|
| FA1 | 21 774 061 | 20 648 522 | 6.2 | 0.03 | 97.79 | 93.3 | 34.16 |
| FA2 | 20 757 300 | 20 132 668 | 6.0 | 0.03 | 97.88 | 93.58 | 36.30 |
| FA3 | 22 134 770 | 21 179 365 | 6.4 | 0.03 | 98.07 | 93.99 | 35.45 |
| MA1 | 21 323 904 | 20 130 023 | 6.0 | 0.03 | 97.82 | 93.43 | 35.49 |
| MA2 | 20 885 385 | 20 454 004 | 6.1 | 0.03 | 97.75 | 93.23 | 33.98 |
| MA3 | 21 288 624 | 20 829 381 | 6.2 | 0.03 | 97.83 | 93.42 | 34.95 |
Tab. 2 Statistics of RNA-Seq of antennae in Monolepta signata
| 样品 Sample | 原始读数 Raw reads | 筛选后读数 Clean reads | 总核酸数(G) Clean bases | 错误率 Error rate (%) | Q20 (%) | Q30 (%) | GC比值 GC content (%) |
|---|---|---|---|---|---|---|---|
| FA1 | 21 774 061 | 20 648 522 | 6.2 | 0.03 | 97.79 | 93.3 | 34.16 |
| FA2 | 20 757 300 | 20 132 668 | 6.0 | 0.03 | 97.88 | 93.58 | 36.30 |
| FA3 | 22 134 770 | 21 179 365 | 6.4 | 0.03 | 98.07 | 93.99 | 35.45 |
| MA1 | 21 323 904 | 20 130 023 | 6.0 | 0.03 | 97.82 | 93.43 | 35.49 |
| MA2 | 20 885 385 | 20 454 004 | 6.1 | 0.03 | 97.75 | 93.23 | 33.98 |
| MA3 | 21 288 624 | 20 829 381 | 6.2 | 0.03 | 97.83 | 93.42 | 34.95 |
| 长度范围 Length range (bp) | Transcripts | Unigenes | ||
|---|---|---|---|---|
| 数量 Number (个) | 百分比 Percentage (%) | 数量 Number (个) | 百分比 Percentage (%) | |
| 300~500 | 53 300 | 33.39 | 29 009 | 39.71 |
| 500~1k | 42 661 | 26.72 | 21 009 | 28.76 |
| 1k~2k | 31 535 | 19.75 | 12 325 | 16.87 |
| >2k | 32 162 | 20.14 | 10 707 | 14.66 |
| 总数Total number | 159 658 | 73 050 | ||
| 总长度Total length | 216 704 872 | 83 267 839 | ||
| 平均长度Mean length | 1 357 | 1 140 | ||
| N50长度N50 length | 2 339 | 1 896 | ||
| N90长度N90 length | 516 | 440 | ||
Tab.3 Summary of the antennal transcriptoms assembly in Monolepta signata
| 长度范围 Length range (bp) | Transcripts | Unigenes | ||
|---|---|---|---|---|
| 数量 Number (个) | 百分比 Percentage (%) | 数量 Number (个) | 百分比 Percentage (%) | |
| 300~500 | 53 300 | 33.39 | 29 009 | 39.71 |
| 500~1k | 42 661 | 26.72 | 21 009 | 28.76 |
| 1k~2k | 31 535 | 19.75 | 12 325 | 16.87 |
| >2k | 32 162 | 20.14 | 10 707 | 14.66 |
| 总数Total number | 159 658 | 73 050 | ||
| 总长度Total length | 216 704 872 | 83 267 839 | ||
| 平均长度Mean length | 1 357 | 1 140 | ||
| N50长度N50 length | 2 339 | 1 896 | ||
| N90长度N90 length | 516 | 440 | ||
| 数据库 Database | 基因数量 Number of unigenes (个) | 百分比 Percentage (%) |
|---|---|---|
| NR | 29 403 | 40.25 |
| NT | 10 676 | 14.61 |
| KEGG | 10 014 | 13.70 |
| Swiss-Prot | 15 487 | 21.20 |
| PFAM | 19 137 | 26.19 |
| GO | 19 135 | 26.19 |
| KOG | 7 554 | 10.34 |
| 所有数据库均有注释 Annotated in all databases | 3 475 | 4.75 |
| 至少有一个数据库注释 Annotated in at least one database | 34 233 | 46.86 |
| 总基因数 Total unigenes | 73 050 |
Tab.4 Successful annotation rate of genes in Monolepta signata
| 数据库 Database | 基因数量 Number of unigenes (个) | 百分比 Percentage (%) |
|---|---|---|
| NR | 29 403 | 40.25 |
| NT | 10 676 | 14.61 |
| KEGG | 10 014 | 13.70 |
| Swiss-Prot | 15 487 | 21.20 |
| PFAM | 19 137 | 26.19 |
| GO | 19 135 | 26.19 |
| KOG | 7 554 | 10.34 |
| 所有数据库均有注释 Annotated in all databases | 3 475 | 4.75 |
| 至少有一个数据库注释 Annotated in at least one database | 34 233 | 46.86 |
| 总基因数 Total unigenes | 73 050 |
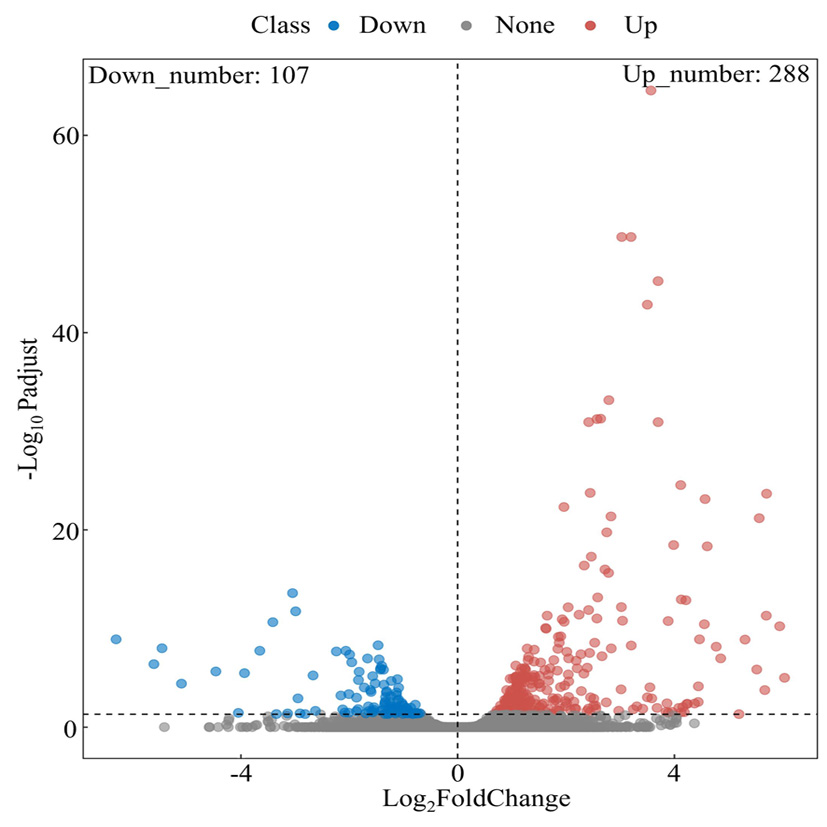
Fig.5 Volcano map of differentially expressed genes Note: The red dots represent up-regulated genes, blue dots down-regulated genes, and gray dots represent genes with no significant differences
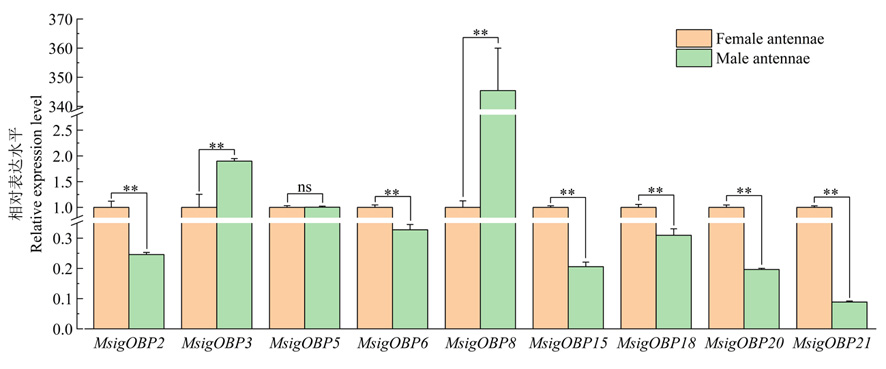
Fig.9 RT-qPCR analysis of odorant binding genes Note: ** Indicates that the difference is extremely significant (P<0.01), * indicates that the difference is significant (P<0.05), ns means no significant difference (P>0.05)
| [1] | 中国科学院动物研究所昆虫分类区系室叶甲组, 河北省张家口地区坝下农业科学研究所植保组, 河北省蔚县农业局植保站西合营公社技术站. 双斑萤叶甲研究简报[J]. 昆虫学报, 1979, 22 (1) : 115-117. |
| Research Group of Leaf Beetle, Division of Insect Taxonomy Institute of Agriculture of Baxia, Changchiakou District, Hopei Province, Technical Station, Plant Protection Station of Xiheying People’s Commune, Agricultural Bureau of Yu County, Hopei Provinge. A preliminary study of the bionomics of the galerucid beetle, Monolepta hieroglyphica (Motschulsky)[J]. Acta Entomologica Sinica, 1979, 22 (1) : 115-117. | |
| [2] | 虞佩玉, 王书永, 杨星科. 中国经济昆虫志,第五十四册,鞘翅目,叶甲总科(二)[M]. 北京: 科学出版社, 1996. |
| YU Peiyu, WANG Shuyong, YANG Xingke. Economic Insect Fauna of China. Coleoptera: Chrysomeloidea (II)[M]. Beijing: Science Press, 1996. | |
| [3] | Wagner T, Bieneck S. Galerucine type material described by victor motschulsky in 1858 and 1866 from the Zoological Museum Moscow (Coleoptera: Chrysomelidae, Galerucinae)[J]. Entomologische Zeitschrift mit Insekten-Börse, 2012, 122 (5) : 205-216. |
| [4] | 梁日霞, 王振营, 何康来, 等. 基于线粒体COⅡ基因序列的双斑长跗萤叶甲中国北方地理种群的遗传多样性研究[J]. 昆虫学报, 2011, 54(7): 828-837. |
| LIANG Rixia, WANG Zhenying, HE Kanglai, et al. Genetic diversity of geographic populations of Monolepta hieroglyphica (Motschulsky) (Coleoptera: Chrysomelidae) from North China estimated by mitochondrial COⅡ gene sequences[J]. Acta Entomologica Sinica, 2011, 54(7): 828-837. | |
| [5] | 陈光辉, 尹弯, 李勤, 等. 双斑长跗萤叶甲研究进展[J]. 中国植保导刊, 2016, 36(10): 19-26. |
| CHEN Guanghui, YIN Wan, LI Qin, et al. Research progress on Monolepta hieroglyphica (Motschulsky)[J]. China Plant Protection, 2016, 36(10): 19-26. | |
| [6] | 李菁, 张小飞, 徐玲玲, 等. 中国南方双斑长跗萤叶甲地理种群遗传结构及Wolbachia感染[J]. 昆虫学报, 2021, 64(6): 730-742. |
| LI Jing, ZHANG Xiaofei, XU Lingling, et al. Genetic structure and Wolbachia infection in geographical populations of Monolepta hieroglyphica(Coleoptera: Chrysomelidae)in South China[J]. Acta Entomologica Sinica, 2021, 64(6): 730-742. | |
| [7] | 赵秀梅, 郑旭, 郭井菲, 等. 齐齐哈尔市玉米田双斑长跗萤叶甲成虫发生规律[J]. 应用昆虫学报, 2021, 58(4): 979-984. |
| ZHAO Xiumei, ZHENG Xu, GUO Jingfei, et al. Occurrence of Monolepta hieroglyphica adults in cornfields in Qiqihar[J]. Chinese Journal of Applied Entomology, 2021, 58(4): 979-984. | |
| [8] | 陈静, 张建萍, 张建华, 等. 双斑长跗萤叶甲的嗜食性[J]. 昆虫知识, 2007, 44(3): 357-360. |
| CHEN Jing, ZHANG Jianping, ZHANG Jianhua, et al. Food preference of Monolepta hieroglyphica[J]. Chinese Bulletin of Entomology, 2007, 44(3): 357-360. | |
| [9] | He Q, Song X M, Ma H W, et al. The complete mitochondrial genome of Monolepta hieroglyphica (Motschulsky) (Coleoptera: Chrysomelidae)[J]. Mitochondrial DNA Part B, 2021, 6(7): 2019-2021.[LinkOut] |
| [10] | 田永浩, 张建萍, 陈静, 等. 新疆棉花新害虫双斑长跗萤叶甲的发生特点及防治策略[J]. 安徽农学通报, 2007, 13(10): 120-121, 230. |
| TIAN Yonghao, ZHANG Jianping, CHEN Jing, et al. Occurring characteristic and preventions and control strategy of Monolepta hieroglyphica(motschulsky)-a new pest of the cotton field in Xinjiang[J]. Anhui Agricultural Science Bulletin, 2007, 13(10): 120-121, 230. | |
| [11] | 杜建军, 云雷. 双斑长跗萤叶甲发生危害特点及防治措施[J]. 陕西农业科学, 2009, 55(3): 202-203. |
| DU Jianjun, YUN Lei. Characteristics and control measures of occurrence and damage of Lepidoptera bimaculata[J]. Shaanxi Journal of Agricultural Sciences, 2009, 55(3): 202-203. | |
| [12] | 李广伟, 陈秀琳. 新疆棉区双斑长跗萤叶甲生活习性及消长动态调查研究[J]. 中国植保导刊, 2010, 30(6): 8-10. |
| LI Guangwei, CHEN Xiulin. Studies on biological characteristics and population dynamics of Monolepta hieroglyphica in cotton in Xinjiang[J]. China Plant Protection, 2010, 30(6): 8-10. | |
| [13] | Zhang X L, Zhang R Y, Li L, et al. Negligible transcriptome and metabolome alterations in RNAi insecticidal maize against Monolepta hieroglyphica[J]. Plant Cell Reports, 2020, 39(11): 1539-1547. |
| [14] |
Zheng F, Jiang H, Jia J L, et al. Effect of dimethoate in controlling Monolepta hieroglyphica (Motschulsky) and its distribution in maize by drip irrigation[J]. Pest Management Science, 2020, 76(4): 1523-1530.
DOI PMID |
| [15] | 戴建青, 韩诗畴, 杜家纬. 植物挥发性信息化学物质在昆虫寄主选择行为中的作用[J]. 环境昆虫学报, 2010, 32(3): 407-414. |
| DAI Jianqing, HAN Shichou, DU Jiawei. Progress in studies on behavioural effect of semiochemicals of host plant to insects[J]. Journal of Environmental Entomology, 2010, 32(3): 407-414. | |
| [16] | 陈秀琳, 李琳琳, 陈玉鑫, 等. 梨小食心虫气味受体GmolOR10基因克隆及表达谱分析[J/OL]. 植物保护: 2023, 1-13 [2023-04-28]. |
| CHEN Xiulin, LI Linlin, CHEN Yuxin, et al. Cloning and expression pattern of odorant receptor GmolOR10 in Grapholita molesta Busck[J/OL]. Plant Protection: 2023, 1-13 [2023-04-28]. | |
| [17] | Pham H T, McNamara K B, Elgar M A. Age-dependent chemical signalling and its consequences for mate attraction in the gumleaf skeletonizer moth, Uraba lugens[J]. Animal Behaviour, 2021, 173: 207-213. |
| [18] | Segura D F, Belliard S A, Vera M T, et al. Plant chemicals and the sexual behavior of male tephritid fruit flies[J]. Annals of the Entomological Society of America, 2018, 111(5): 239-264. |
| [19] | 赵航, 吴国星, 汤永玉, 等. 叉角厉蝽触角转录组及嗅觉相关基因分析[J]. 环境昆虫学报, 2022, 44(5): 1205-1217. |
| ZHAO Hang, WU Guoxing, TANG Yongyu, et al. Analysis of the antennal transcriptome and olfaction-related genes of Eocanthecona furcellata[J]. Journal of Environmental Entomology, 2022, 44(5): 1205-1217. | |
| [20] | 张聪, 王振营, 何康来, 等. 双斑长跗萤叶甲触角感器的扫描电镜观察[J]. 应用昆虫学报, 2012, 49(3): 756-761. |
| ZHANG Cong, WANG Zhenying, HE Kanglai, et al. Scanning electron microscopy studies of antennal sensilla of Monolepta hieroglyphica[J]. Chinese Journal of Applied Entomology, 2012, 49(3): 756-761. | |
| [21] | 刘敏, 刘爱萍, 韩海斌. 寄生蜂转录组学研究进展[J]. 植物保护, 2021, 47(2): 11-19. |
| LIU Min, LIU Aiping, HAN Haibin. A review of transcriptome sequencing in parasitoid wasps[J]. Plant Protection, 2021, 47(2): 11-19. | |
| [22] |
Güell M, van Noort V, Yus E, et al. Transcriptome complexity in a genome-reduced bacterium[J]. Science, 2009, 326(5957): 1268-1271.
DOI PMID |
| [23] | 刘伟, 郭光艳, 秘彩莉. 转录组学主要研究技术及其应用概述[J]. 生物学教学, 2019, 44(10): 2-5. |
| LIU Wei, GUO Guangyan, BEI Caili. A summary of the main research techniques of transcriptology and their applications[J]. Biology Teaching, 2019, 44(10): 2-5. | |
| [24] | 李资聪, 刘磊, 杨斌, 等. 黄野螟化学感受基因的鉴定与分析[J]. 植物保护, 2021, 47(6): 34-48. |
| LI Zicong, LIU Lei, YANG Bin, et al. Identification and analysis of chemosensory genes in Heortia vitessoides[J]. Plant Protection, 2021, 47(6): 34-48. | |
| [25] | 白鹏华, 王冰, 张仙红, 等. 昆虫气味受体的研究方法与进展[J]. 昆虫学报, 2022, 65(3): 1-22. |
| BAI Penghua, WANG Bing, ZHANG Xianhong, et al. Research methods and advances of odorant receptors in insects[J]. Acta Entomologica Sinica, 2022, 65(3): 1-22. | |
| [26] |
詹文会, 张苏芳, 耿红卫, 等. 天牛嗅觉感受相关蛋白研究进展[J]. 河南农业科学, 2018, 47(3): 1-6.
DOI |
| ZHAN Wenhui, ZHANG Sufang, GENG Hongwei, et al. Research progress on olfactory recognition proteins of longhorn beetle[J]. Journal of Henan Agricultural Sciences, 2018, 47(3): 1-6. | |
| [27] | Rondoni G, Roman A, Meslin C, et al. Antennal transcriptome analysis and identification of candidate chemosensory genes of the harlequin ladybird beetle, Harmonia axyridis (Pallas) (Coleoptera: Coccinellidae)[J]. Insects, 2021, 12(3): 209. |
| [28] | 庄翔麟, 吉帅帅, 赵昱杰, 等. 灭字脊虎天牛CSP基因的鉴定、表达谱及XquaCSP7基因的功能研究[J]. 植物保护, 2020, 46(3): 30-39. |
| ZHUANG Xianglin, JI Shuaishuai, ZHAO Yujie, et al. Identification and expression pattern of chemosensory protein genes from Xylotrechus quadripes Chevrolat (Coleoptera: Cerambycidae) and functional studies of XquaCSP7 gene[J]. Plant Protection, 2020, 46(3): 30-39. | |
| [29] | Wang X, Wang S, Yi J K, et al. Three host plant volatiles, hexanal, lauric acid, and tetradecane, are detected by an antenna-biased expressed odorant receptor 27 in the dark black chafer Holotrichia parallela[J]. Journal of Agricultural and Food Chemistry, 2020, 68(28): 7316-7323. |
| [30] | Yuvaraj J K, Roberts R E, Sonntag Y, et al. Putative ligand binding sites of two functionally characterized bark beetle odorant receptors[J]. BMC Biology, 2021, 19(1): 16. |
| [31] | Hong B, Chang Q, Zhai Y J, et al. Functional characterization of odorant binding protein PyasOBP2 from the jujube bud weevil, Pachyrhinus yasumatsui (Coleoptera: Curculionidae) 2022. |
| [32] |
Grabherr M G, Haas B J, Yassour M, et al. Full-length transcriptome assembly from RNA-Seq data without a reference genome[J]. Nature Biotechnology, 2011, 29(7): 644-652.
DOI PMID |
| [33] | Zhang Y N, Kang K, Xu L, et al. Deep sequencing of antennal transcriptome from Callosobruchus chinensis to characterize odorant binding protein and chemosensory protein genes[J]. Journal of Stored Products Research, 2017, 74 : 13-21. |
| [34] | 黄聪, 李有志, 杨念婉, 等. 入侵昆虫基因组研究进展[J]. 植物保护, 2019, 45(5): 112-120, 134. |
| HUANG Cong, LI Youzhi, YANG Nianwan, et al. Progresses in invasive insect genomics[J]. Plant Protection, 2019, 45(5): 112-120, 134. | |
| [35] | Gu X C, Zhang Y N, Kang K, et al. Antennal transcriptome analysis of odorant reception genes in the red turpentine beetle (RTB), Dendroctonus valens[J]. PLoS One, 2015, 10(5): e0125159.[PubMed] |
| [36] | 杨雪娇, 张蔓, 江宇航, 等. 七星瓢虫触角转录组及嗅觉相关基因分析[J]. 昆虫学报, 2020, 63(6): 717-726. |
| YANG Xuejiao, ZHANG Man, JIANG Yuhang, et al. Analysis of the antennal transcriptome and olfaction-related genes of Coccinella septempunctata (Coleoptera: Coccinellidae)[J]. Acta Entomologica Sinica, 2020, 63(6): 717-726. | |
| [37] | 巩雪芳, 谢寿安, 杨平, 等. 花椒窄吉丁触角转录组及嗅觉相关基因分析[J]. 昆虫学报, 2020, 63(10): 1159-1170. |
| GONG Xuefang, XIE Shouan, YANG Ping, et al. Analysis of the antennal transcriptome and olfaction-related genes of Agrilus zanthoxylumi (Coleoptera: Buprestidae)[J]. Acta Entomologica Sinica, 2020, 63(10): 1159-1170. | |
| [38] | 胡佳萌, 徐丹萍, 卓志航, 等. 云斑天牛成虫触角转录组及嗅觉相关基因分析[J]. 应用昆虫学报, 2019, 56(5): 1037-1047. |
| HU Jiameng, XU Danping, ZHUO Zhihang, et al. Analysis of antennal transcriptome and olfaction-related genes of adult Batocera horsfieldi (Hope)[J]. Chinese Journal of Applied Entomology, 2019, 56(5): 1037-1047. | |
| [39] |
宋月芹, 董钧锋, 陈庆霄, 等. 点蜂缘蝽触角转录组及化学感受相关基因的分析[J]. 昆虫学报, 2017, 60(10): 1120-1128.
DOI |
|
SONG Yueqin, DONG Junfeng, CHEN Qingxiao, et al. Analysis of the antennal transcriptome and chemoreception-related genes of the bean bug, Riptortus pedestris (Hemiptera: Alydidae)[J]. Acta Entomologica Sinica, 2017, 60(10): 1120-1128.
DOI |
|
| [40] | 张健, 谢婉莹, 董曼羽, 等. 青杨天牛嗅觉相关基因的鉴定及表达谱分析[J]. 江苏农业科学, 2022, 50(4): 23-28. |
| ZHANG Jian, XIE Wanying, DONG Manyu, et al. Identification and expression profile analysis of olfactory related genes of Anoplophora populnea[J]. Jiangsu Agricultural Sciences, 2022, 50(4): 23-28. | |
| [41] | 龙亚芹, 罗梓文, 王雪松, 等. 茶谷蛾成虫触角转录组及嗅觉相关基因分析[J]. 茶叶科学, 2021, 41(4): 553-563. |
| LONG Yaqin, LUO Ziwen, WANG Xuesong, et al. Analysis of the antennal transcriptome and olfactory-related genes in the Agriophara rhombata[J]. Journal of Tea Science, 2021, 41(4): 553-563. | |
| [42] | 杨爽, 赵慧婷, 徐凯, 等. 大蜡螟触角转录组及嗅觉相关基因分析[J]. 应用昆虫学报, 2019, 56(6): 1279-1291. |
| YANG Shuang, ZHAO Huiting, XU Kai, et al. Analysis of the antennal transcriptome and olfaction-related genes of the greater wax moth Galleria mellonella(Lepidoptera: Pyralidae)[J]. Chinese Journal of Applied Entomology, 2019, 56(6): 1279-1291. | |
| [43] | Wu G X, Su R R, Ouyang H L, et al. Antennal transcriptome analysis and identification of olfactory genes in Glenea cantor Fabricius (Cerambycidae: Lamiinae)[J]. Insects, 2022, 13(6): 553. |
| [44] | 巩雪芳, 谢寿安, 杨平, 等. 花椒窄吉丁雌、雄成虫触角差异表达基因的初步筛选[J]. 农业生物技术学报, 2019, 27(7): 1233-1245. |
| GONG Xuefang, XIE Shouan, YANG Ping, et al. Preliminary screening of differentially expressed genes of female and male antenna of Agrilus zanthoxylumi[J]. Journal of Agricultural Biotechnology, 2019, 27(7): 1233-1245. | |
| [45] | Tanaka K, Shimomura K, Hosoi A, et al. Antennal transcriptome analysis of chemosensory genes in the cowpea beetle, Callosobruchus maculatus (F.)[J]. PLoS One, 2022, 17(1): e0262817. |
| [1] | MENG Hanying, GUO Dandan, HE Wanjie, ZHANG Weiwei, ZHANG Jianping, CHEN Jing. Effects of different feeding time of Monolepta Signata on the composition and content of volatile in maize leaves [J]. Xinjiang Agricultural Sciences, 2024, 61(2): 421-433. |
| [2] | WANG Wei, ZHANG Renfu, LIU Haiyang, DING Ruifeng, LIANG Gemei, YAO Ju. Transcriptome analysis of Aphis gossypii sulfoxaflor and acetamiprid-resistant strains with different genetic backgrounds of resistance [J]. Xinjiang Agricultural Sciences, 2024, 61(12): 3078-3088. |
| [3] | SONG Jikun, XIN Yue, LI Longyun, LIU Guoyuan, PEI Wenfeng, MA Jianjiang, QU Yanying, YU Jiwen, WU Man. Comparative Analysis of Gene Expression between Giza 75 and SG 747 Using Cotton Oligonucleotide Microarrays during Fiber Development [J]. Xinjiang Agricultural Sciences, 2022, 59(6): 1312-1330. |
| [4] | MA Meng-ting;WANG Shan-shan;CHEN Xiao-li;Yisila rmu;GAO Qing-hua;HAN Chun-mei. Differentially expressed genes in X-and Y-sperm from bovine screened by subtractive hybridization [J]. , 2015, 52(4): 774-778. |
| Viewed | ||||||||||||||||||||||||||||||||||||||||||||||||||
|
Full text 44
|
|
|||||||||||||||||||||||||||||||||||||||||||||||||
|
Abstract 124
|
|
|||||||||||||||||||||||||||||||||||||||||||||||||