

Xinjiang Agricultural Sciences ›› 2024, Vol. 61 ›› Issue (6): 1301-1309.DOI: 10.6048/j.issn.1001-4330.2024.06.001
• Crop Genetics and Breeding • Germplasm Resources?Molecular Genetics • Cultivation Physiology • Physiology and Biochemistry • Previous Articles Next Articles
GONG Junming( ), XIONG Xianpeng, ZHANG Caixia, SHAO Dongnan, CHENG Shuaishuai, SUN Jie(
), XIONG Xianpeng, ZHANG Caixia, SHAO Dongnan, CHENG Shuaishuai, SUN Jie( )
)
Received:2023-10-23
Online:2024-06-20
Published:2024-08-08
Correspondence author:
SUN Jie
Supported by:
巩隽铭( ), 熊显鹏, 张彩霞, 邵东南, 程帅帅, 孙杰(
), 熊显鹏, 张彩霞, 邵东南, 程帅帅, 孙杰( )
)
通讯作者:
孙杰
作者简介:巩隽铭(1995- ),男,辽宁人,硕士研究生,研究方向为棉花分子育种,(E-mail) gongjunming_cn@163.com
基金资助:CLC Number:
GONG Junming, XIONG Xianpeng, ZHANG Caixia, SHAO Dongnan, CHENG Shuaishuai, SUN Jie. Functional analysis of 4-coumarate: CoA ligase gene Gh4CL30 in upland cotton[J]. Xinjiang Agricultural Sciences, 2024, 61(6): 1301-1309.
巩隽铭, 熊显鹏, 张彩霞, 邵东南, 程帅帅, 孙杰. 陆地棉4-香豆酸辅酶A连接酶基因Gh4CL30的功能分析[J]. 新疆农业科学, 2024, 61(6): 1301-1309.
| 基因名称 Genes Name | 基因ID Gene ID | 染色体 Chromosome | 蛋白长度 Protein Length (aa) | 等电点 Isoelectric Point | 亚细胞定位 Subcellular localization |
|---|---|---|---|---|---|
| Gh4CL6 | GH_A05G0051 | A05 | 543 | 5.835 | Plasma membrane |
| Gh4CL7 | GH_A05G1439 | A05 | 543 | 6.09 | cytosol |
| Gh4CL12 | GH_A10G0499 | A09 | 129 | 10.388 | chloroplast |
| Gh4CL24 | GH_D05G0056 | D05 | 540 | 6.25 | chloroplast |
| Gh4CL25 | GH_D05G1456 | D05 | 543 | 5.872 | cytosol |
| Gh4CL30 | GH_D10G0525 | D10 | 543 | 5.284 | cytosol |
Tab.1 Type I Gh4CL genes in G.hirsutum
| 基因名称 Genes Name | 基因ID Gene ID | 染色体 Chromosome | 蛋白长度 Protein Length (aa) | 等电点 Isoelectric Point | 亚细胞定位 Subcellular localization |
|---|---|---|---|---|---|
| Gh4CL6 | GH_A05G0051 | A05 | 543 | 5.835 | Plasma membrane |
| Gh4CL7 | GH_A05G1439 | A05 | 543 | 6.09 | cytosol |
| Gh4CL12 | GH_A10G0499 | A09 | 129 | 10.388 | chloroplast |
| Gh4CL24 | GH_D05G0056 | D05 | 540 | 6.25 | chloroplast |
| Gh4CL25 | GH_D05G1456 | D05 | 543 | 5.872 | cytosol |
| Gh4CL30 | GH_D10G0525 | D10 | 543 | 5.284 | cytosol |
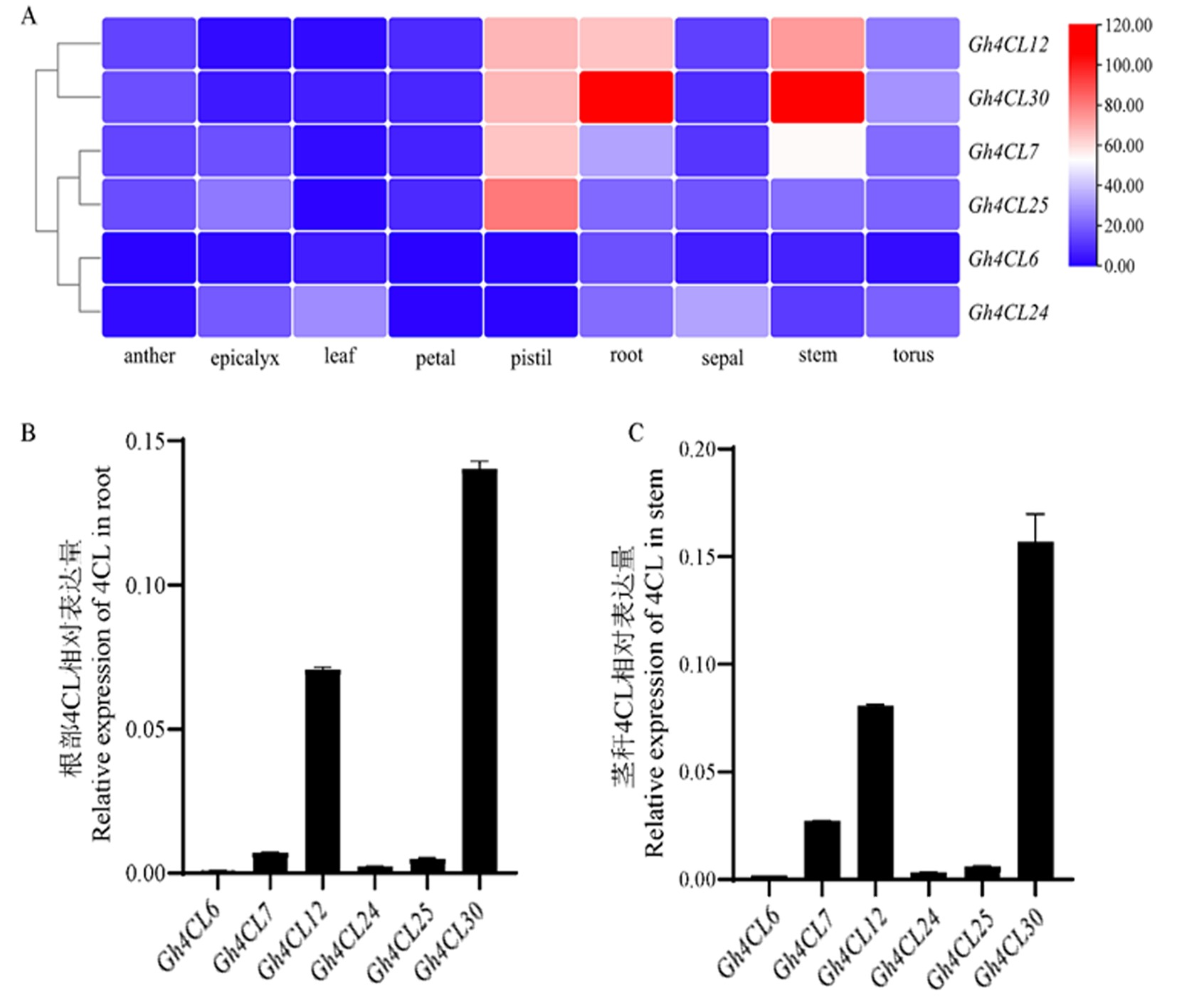
Fig.1 Expression patterns of six type I Gh4CL in different tissues of cotton Note: A:Expression patterns of six type I Gh4CLs in different tissues of cotton;B:Relative expression of six type I Gh4CL genes in cotton roots;C:Relative expression of six type I Gh4CL genes in cotton stems
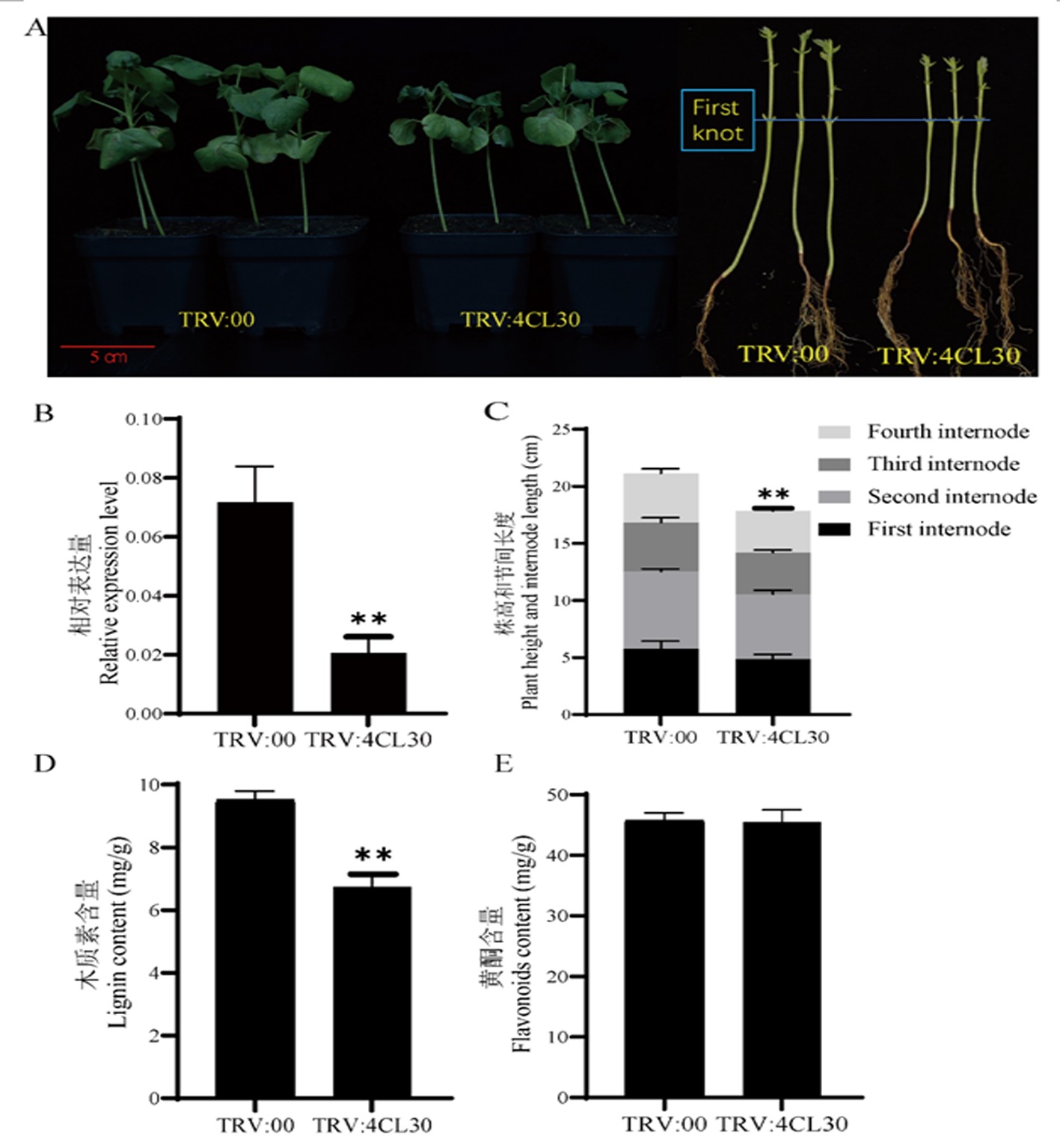
Fig.2 Phenotype and determination of relevant indicators in Gh4CL30 gene silenced plants Note: A:Phenotypic differences between TRV:Gh4CL30 and TRV:00 plants;B:Relative expression of Gh4CL30 in TRV:00 and TRV:Gh4CL30 plants;C:Differences in plant height and internode length between TRV:00 and TRV:Gh4CL30 plants.From bottom to top, the first to fourth internode lengths of cotton plants are shown;D:Lignin content of TRV:00 and TRV:Gh4CL30 plants;E:Flavonoid metabolite content of TRV:00 and TRV:Gh4CL30 plants
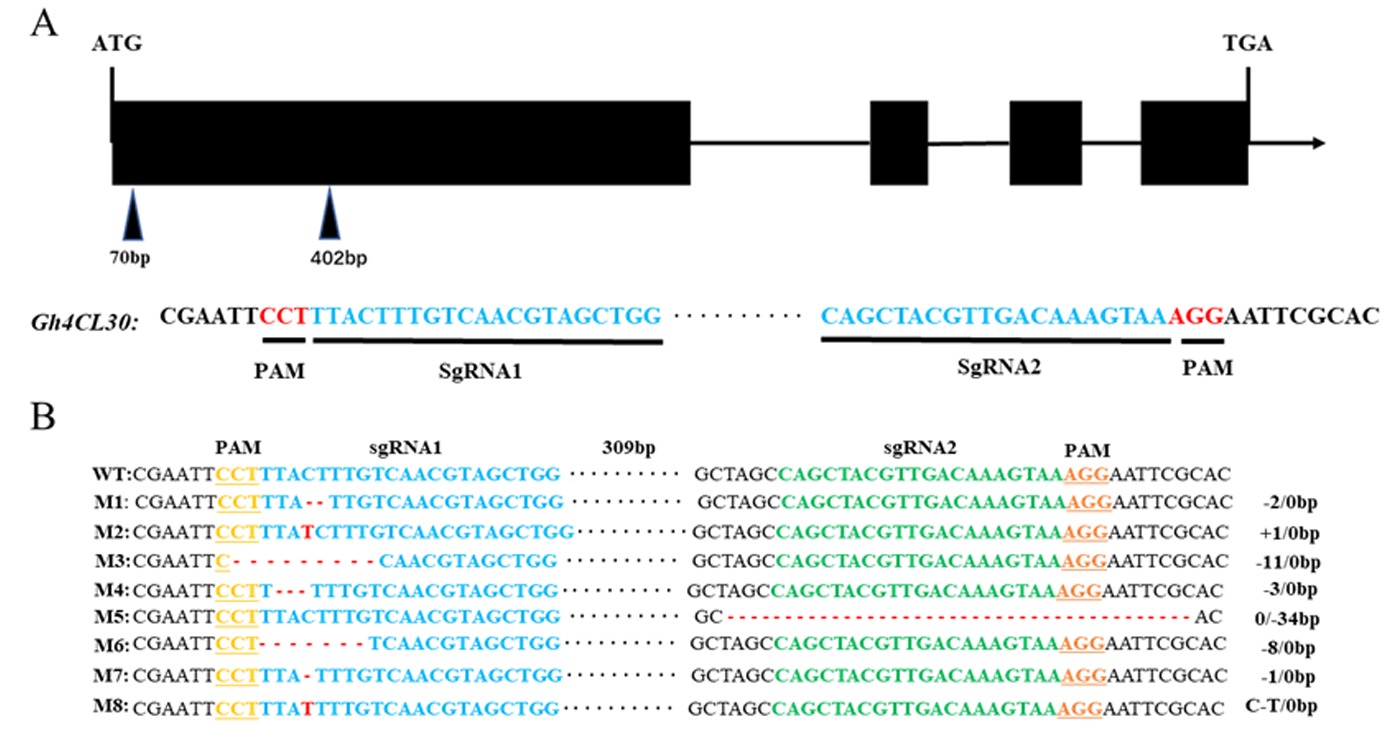
Fig.3 Gh4CL30 gene structure and selection of editing targets and types of gene editing Note:A:Two editing targets were selected in the first exon region of the Gh4CL30 gene;B:Type of gene editing of the mutant
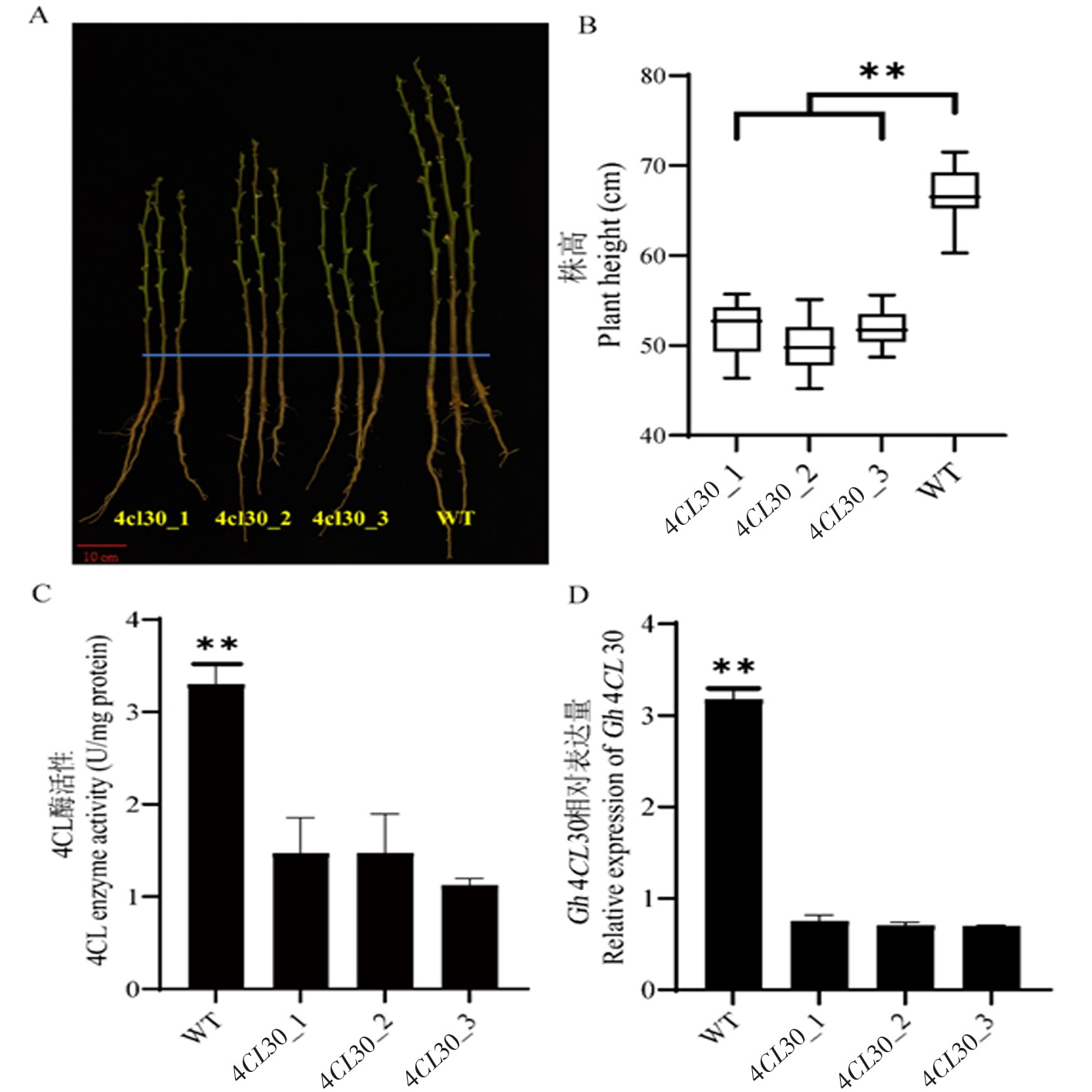
Fig.4 Phenotype, 4CL enzyme activity and relative expression of Gh4CL30 in WT and 4CL30 plants Note: A:Phenotypic comparison of 4CL30 and WT plant heights;B:Plant height statistics of 4CL30 edited plants and WT; C:The 4CL enzyme activity of WT and 4CL30 plants was different;D:Relative expression of Gh4CL30 gene in stems of WT and 4CL30 edited plants
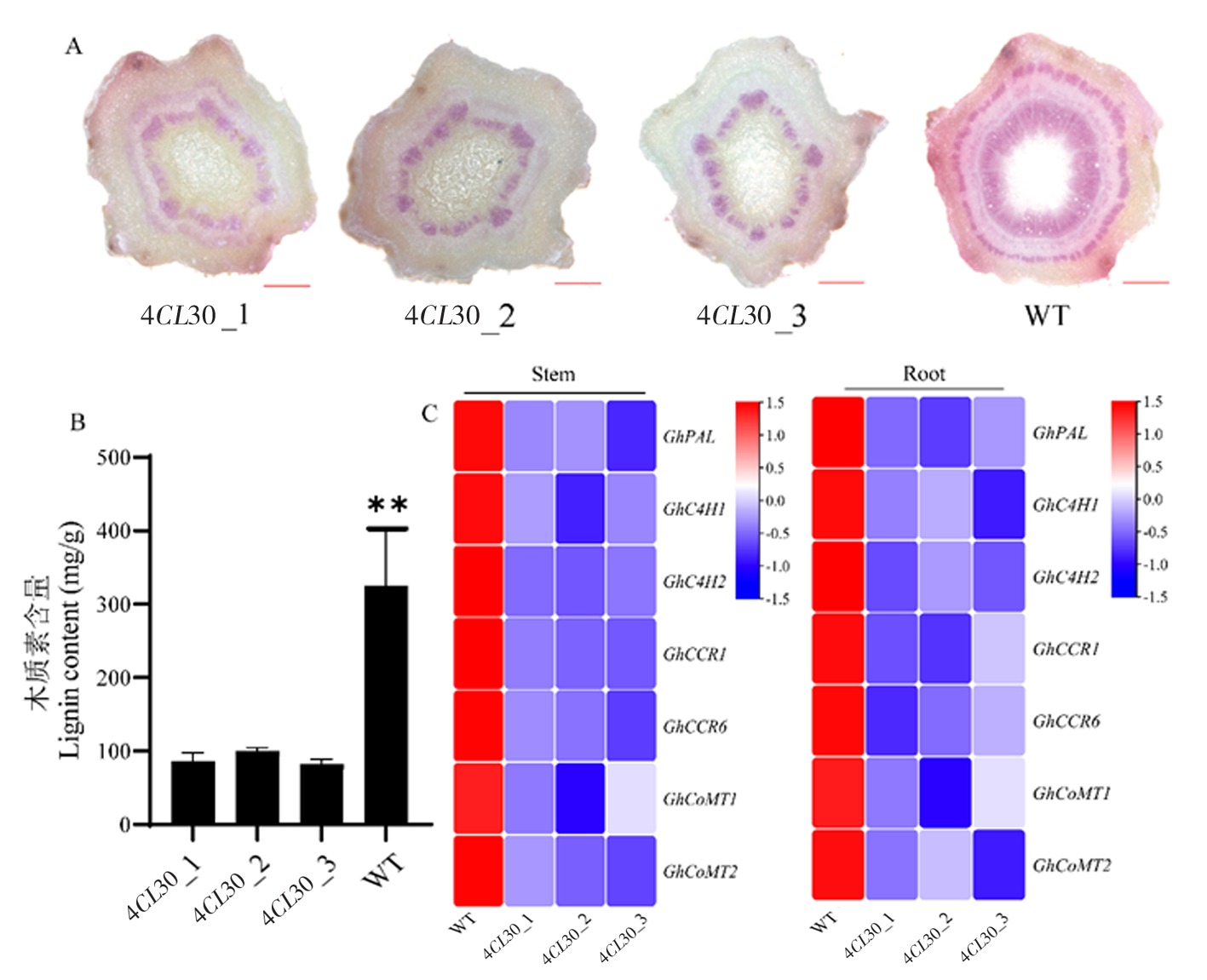
Fig.5 Lignin content of 4CL30 and WT plants and expression of genes related to lignin synthesis Note:A:Staining of stem sections of 4CL30 and WT plants at the same node using hydrochloric acid-phloroglucinol;B:Determination of lignin content in 4CL30 and WT plants;C:Expression of genes related to lignin synthesis in stem and root tissues of 4CL30 and WT plants
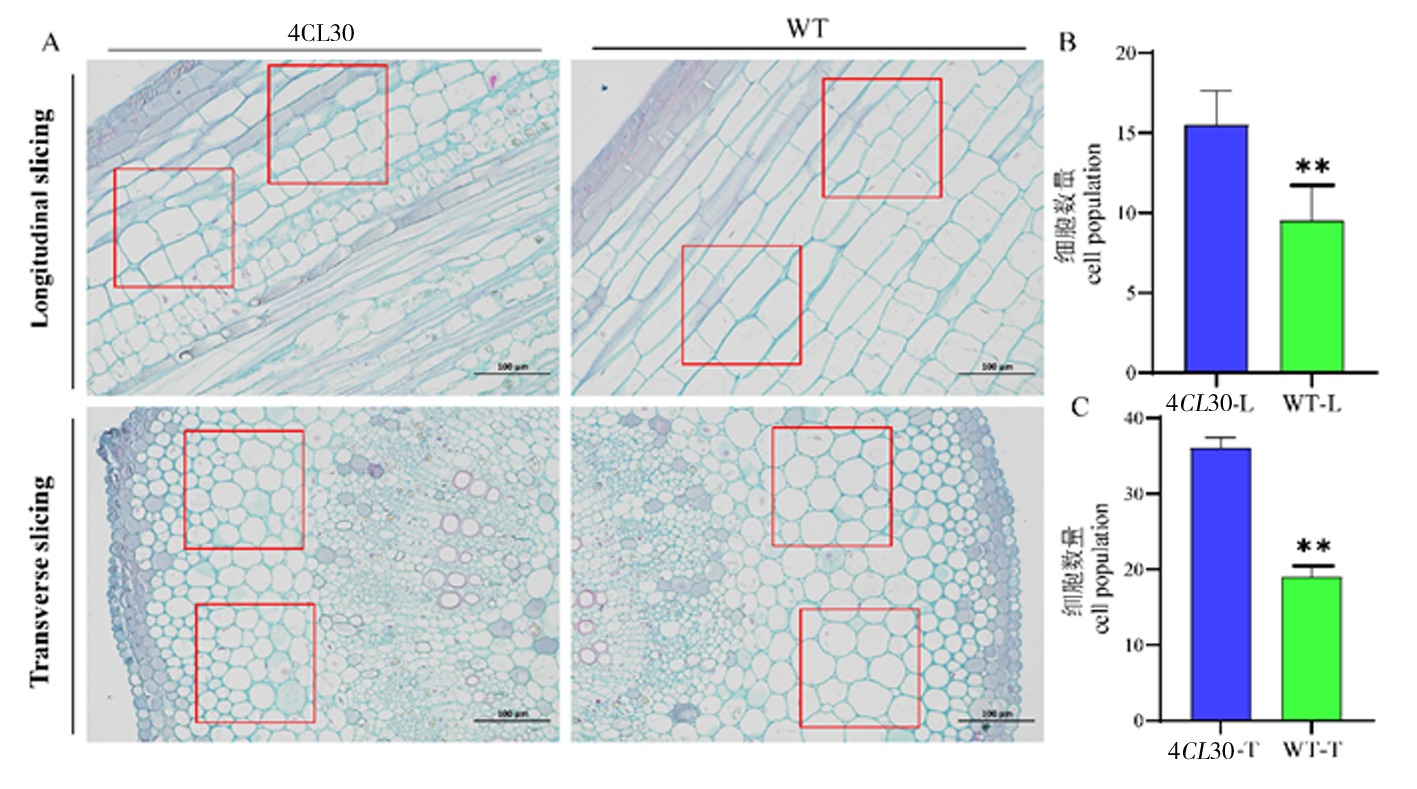
Fig.6 Cytological observations of 4CL30 and WT plants Note:A:Transverse and longitudinal paraffin sections of same-node stems of 4CL30 and WT plants;B:Cell counts in longitudinal sections;C:Cell counting in transverse sections (Cells in red boxes of the same size are counted at the same scale; a small number of cells indicates a longer cell length and a larger cell size.)
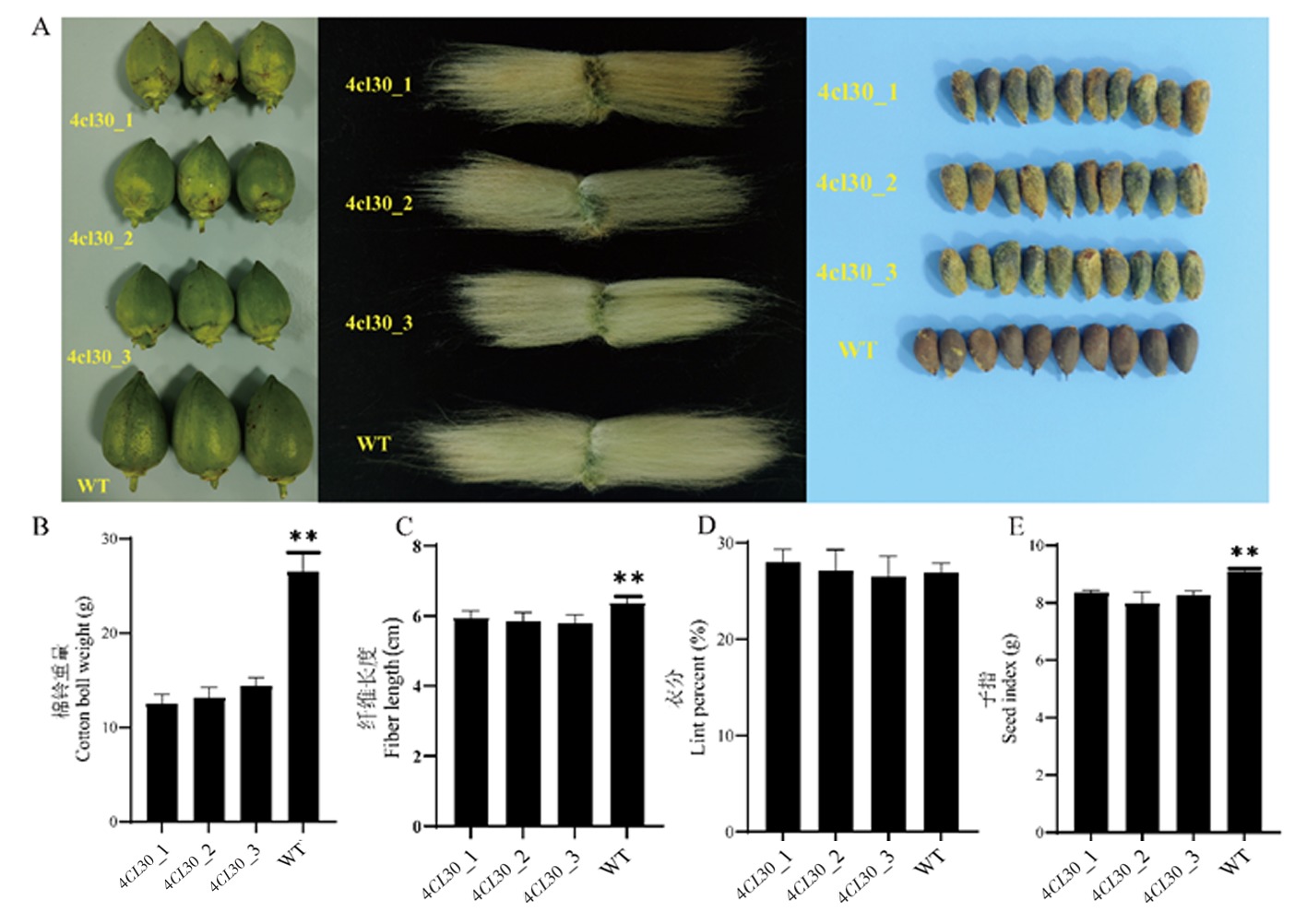
Fig.7 Phenotypic comparison of boll size and fiber length of WT and 4CL30 plants Note:A:From left to right are plant boll size, fiber length and seed size;B:Cotton boll weight (Cotton bolls on the first fruit branch of the same period) of WT and 4CL30 plants;C:Fiber length of WT and 4CL30 plants;D:Lint percent of WT and 4CL30 plants;E:Seed index of WT and 4CL30 plants
| [1] | Sun Q, Xie Y H, Li H M, et al. Cotton GhBRC1 regulates branching, flowering, and growth by integrating multiple hormone pathways[J]. The Crop Journal, 2022, 10(1): 75-87. |
| [2] | Bensen R J, Johal G S, Crane V C, et al. Cloning and characterization of the maize An1 gene[J]. The Plant Cell, 1995, 7(1): 75-84. |
| [3] |
Fu J Y, Ren F, Lu X, et al. A tandem array of ent-kaurene synthases in maize with roles in gibberellin and more specialized metabolism[J]. Plant Physiology, 2016, 170(2): 742-751.
DOI PMID |
| [4] |
Vogt T. Phenylpropanoid biosynthesis[J]. Molecular Plant, 2010, 3(1): 2-20.
DOI PMID |
| [5] |
Wang S C, Alseekh S, Fernie A R, et al. The structure and function of major plant metabolite modifications[J]. Molecular Plant, 2019, 12(7): 899-919.
DOI PMID |
| [6] |
Zhao Q. Lignification: flexibility, biosynthesis and regulation[J]. Trends in Plant Science, 2016, 21(8): 713-721.
DOI PMID |
| [7] |
Nakabayashi R, Saito K. Integrated metabolomics for abiotic stress responses in plants[J]. Current Opinion in Plant Biology, 2015, 24: 10-16.
DOI PMID |
| [8] |
Le Roy J, Huss B, Creach A, et al. Glycosylation is a major regulator of phenylpropanoid availability and biological activity in plants[J]. Frontiers in Plant Science, 2016, 7: 735.
DOI PMID |
| [9] |
Amrhein N, Frank G, Lemm G, et al. Inhibition of lignin formation by L-alpha-aminooxy-beta-phenylpropionic acid, an inhibitor of phenylalanine ammonia-lyase[J]. European Journal of Cell Biology, 1983, 29(2): 139-144.
PMID |
| [10] | Smart C C, Amrhein N. The influence of lignification on the development of vascular tissue inVigna radiata L[J]. Protoplasma, 1985, 124(1): 87-95. |
| [11] |
Lavhale S G, Kalunke R M, Giri A P. Structural, functional and evolutionary diversity of 4-coumarate-CoA ligase in plants[J]. Planta, 2018, 248(5): 1063-1078.
DOI PMID |
| [12] | Voelker S L, Lachenbruch B, Meinzer F C, et al. Reduced wood stiffness and strength, and altered stem form, in young antisense 4CL transgenic poplars with reduced lignin contents[J]. The New Phytologist, 2011, 189(4): 1096-1109. |
| [13] | Chen X H, Wang H T, Li X Y, et al. Molecular cloning and functional analysis of 4-Coumarate: CoA ligase 4(4CL-like 1)from Fraxinus mandshurica and its role in abiotic stress tolerance and cell wall synthesis[J]. BMC Plant Biology, 2019, 19(1): 231. |
| [14] | Li Y, Kim J I, Pysh L, et al. Four isoforms of Arabidopsis 4-coumarate: CoA ligase have overlapping yet distinct roles in phenylpropanoid metabolism[J]. Plant Physiology, 2015, 169(4): 2409-2421. |
| [15] | Gui J S, Shen J H, Li L G. Functional characterization of evolutionarily divergent 4-coumarate: coenzyme a ligases in rice[J]. Plant Physiology, 2011, 157(2): 574-586. |
| [16] | Shi R, Sun Y H, Li Q Z, et al. Towards a systems approach for lignin biosynthesis in Populus trichocarpa: transcript abundance and specificity of the monolignol biosynthetic genes[J]. Plant & Cell Physiology, 2010, 51(1): 144-163. |
| [17] | Ehlting J, Büttner D, Wang Q, et al. Three 4-coumarate: coenzyme A ligases in Arabidopsis thaliana represent two evolutionarily divergent classes in angiosperms[J]. The Plant Journal, 1999, 19(1): 9-20. |
| [18] |
Wang B, Sun W, Li Q S, et al. Genome-wide identification of phenolic acid biosynthetic genes in Salvia miltiorrhiza[J]. Planta, 2015, 241(3): 711-725.
DOI PMID |
| [19] | Sun S C, Xiong X P, Zhang X L, et al. Characterization of the Gh4CL gene family reveals a role of Gh4CL7 in drought tolerance[J]. BMC Plant Biology, 2020, 20(1): 125. |
| [20] | Zhang T Z, Hu Y, Jiang W K, et al. Sequencing of allotetraploid cotton (Gossypium hirsutum L. acc. TM-1) provides a resource for fiber improvement[J]. Nature Biotechnology, 2015, 33(5): 531-537. |
| [21] |
Chen C J, Chen H, Zhang Y, et al. TBtools: an integrative toolkit developed for interactive analyses of big biological data[J]. Molecular Plant, 2020, 13(8): 1194-1202.
DOI PMID |
| [22] | Gao W, Long L, Zhu L F, et al. Proteomic and virus-induced gene silencing (VIGS) Analyses reveal that gossypol, brassinosteroids, and jasmonic acid contribute to the resistance of cotton to Verticillium dahliae[J]. Molecular & Cellular Proteomics, 2013, 12(12): 3690-3703. |
| [23] | Cheng X Q, Zhang X Y, Xue F, et al. Characterization and transcriptome analysis of a dominant genic male sterile cotton mutant[J]. BMC Plant Biology, 2020, 20(1): 312. |
| [24] | Fan L, Shi W J, Hu W R, et al. Molecular and biochemical evidence for phenylpropanoid synthesis and presence of wall-linked phenolics in cotton fibers[J]. Journal of Integrative Plant Biology, 2009, 51(7): 626-637. |
| [25] | 付远志, 薛惠云, 胡根海, 等. 我国棉花株型性状遗传育种研究进展[J]. 江苏农业科学, 2019, 47(5): 16-19. |
| FU Yuanzhi, XUE Huiyun, HU Genhai, et al. Research progress on genetics breeding of plant architecture traits of cotton in China[J]. Jiangsu Agricultural Sciences, 2019, 47(5): 16-19. | |
| [26] | Li X, Bonawitz N D, Weng J K, et al. The growth reduction associated with repressed lignin biosynthesis in Arabidopsis thaliana is independent of flavonoids[J]. The Plant Cell, 2010, 22(5): 1620-1632. |
| [27] |
Hu W J, Kawaoka A, Tsai C J, et al. Compartmentalized expression of two structurally and functionally distinct 4-coumarate: CoA ligase genes in aspen (Populus tremuloides)[J]. Proceedings of the National Academy of Sciences of the United States of America, 1998, 95(9): 5407-5412.
DOI PMID |
| [28] | Sutela S, Hahl T, Tiimonen H, et al. Phenolic compounds and expression of 4CL genes in silver birch clones and Pt4CL1a lines[J]. PLoS One, 2014, 9(12): e114434. |
| [29] |
Allina S M, Pri-Hadash A, Theilmann D A, et al. 4-Coumarate: coenzyme A ligase in hybrid poplar. Properties of native enzymes, cDNA cloning, and analysis of recombinant enzymes[J]. Plant Physiology, 1998, 116(2): 743-754.
PMID |
| [30] | Zhong R Q, Cui D T, Ye Z H. Secondary cell wall biosynthesis[J]. The New Phytologist, 2019, 221(4): 1703-1723. |
| [31] | Gou J Y, Wang L J, Chen S P, et al. Gene expression and metabolite profiles of cotton fiber during cell elongation and secondary cell wall synthesis[J]. Cell Research, 2007, 17(5): 422-434. |
| [1] | WANG Kaidi, GAO Chenxu, PEI Wenfeng, YANG Shuxian, ZHANG Wenqing, SONG Jikun, MA Jianjiang, WANG Li, YU Jiwen, CHEN Quanjia. Identification of TRM gene family and fiber quality related excellent haplotype analysis in Gossypium hirsutum L. [J]. Xinjiang Agricultural Sciences, 2024, 61(3): 521-536. |
| [2] | GENG Feifei, MENG Chaomin, QING Guixia, ZHOU Jiamin, ZHANG Fuhou, LIU Fengju. Cloning and expression analysis of phosphorus efficient gene GhMYB4 in Gossypium hirsutum L. [J]. Xinjiang Agricultural Sciences, 2023, 60(6): 1406-1412. |
| [3] | LI Peiqi, SUN Qingpei, WANG Zhihui, QIN Xinzheng, FAN Yonghong. Correlation analysis of lignin degradation and enzyme activity changes in solid fermentation of cotton stalks [J]. Xinjiang Agricultural Sciences, 2023, 60(6): 1423-1432. |
| [4] | WEN Jia, HUANG Chenjue, JI Zihan, LI Libei, FENG Zhen, YU Shuxun. Association analysis of dynamic plant height trait using SSR marker in Gossypium hirsutum L. [J]. Xinjiang Agricultural Sciences, 2023, 60(12): 2892-2901. |
| [5] | GAO Qian, WANG Yamei, WU Pingfan, ZHANG Hongmei, ZHOU Ling. Determination of Fiber Component Content in the Residual Branches of Fruit Trees in South Xinjiang Based on Near Infrared Spectroscopy [J]. Xinjiang Agricultural Sciences, 2022, 59(8): 2025-2032. |
| [6] | PAN Zhiyuan, DUAN Yanyan, CHANG Baoxue, CUI Zhenkun, QI Bingqin, HU Jing, GOU Ling. Research on the Lodging Resistance Characteristics and Heterosis in Different Height Maize Hybrids [J]. Xinjiang Agricultural Sciences, 2022, 59(7): 1590-1597. |
| [7] | YANG Bo, CHE Yuhong, GUO Chunmiao, Mubareke·Ayoupu, WU Jingrong, DU Juan. Cloning and bioinformation analysis of 4CL gene in Cydonia oblonga Mill [J]. Xinjiang Agricultural Sciences, 2021, 58(6): 1055-1063. |
| [8] | Omarjan Kurban, Nusrat Osiman, Arman Ablimet, LI Jie, ZHANG Pengzhong, Tursunjan Mamat. Comprehensive Evaluation and Analysis of Main Characters of Different Cotton Cultivars in Oasis Arid Area in Xinjiang [J]. Xinjiang Agricultural Sciences, 2021, 58(4): 643-652. |
| [9] | ZHANG Yi, SHAO Dongnan, XUE Fei, LI Huqing, WANG Xuefeng, LI Yanjun, ZHANG Xinyu, LIU Feng, SUN Jie. Cloning and Functional Analysis of Cytochrome P450 Superfamily Gene GhCYP85A2-1 in Upland Cotton [J]. Xinjiang Agricultural Sciences, 2021, 58(2): 197-205. |
| [10] | JIN Yinan, DONG Helin, LI Pengcheng, SUN Miao, SHAO Jingjing, FENG Weina, XU Wenxiu, ZHENG Cangsong. Effects of soil potassium level on growth and photosynthetic characteristics of early cotton [J]. Xinjiang Agricultural Sciences, 2021, 58(12): 2236-2243. |
| [11] | ZHANG Yan, YAO Yinkun, HU We, GAO Yuan, TANG Mingyao. Effects of Phosphate Fertilizer Application on P Accumulation, Distribution,Utilization and Yield of Cotton [J]. Xinjiang Agricultural Sciences, 2020, 57(11): 2004-2011. |
| [12] | CHEN Xiang-dong, WU Xiao-jun, HU Xi-gui, LI Gan, JIANG Xiao-ling, LI Xiao-hui, LI Xiao-li, HU Tie-zhu, RU Zhen-gang. Molecular Detection of Five Dwarf Genes in 117 Wheat Germplasm Resources [J]. Xinjiang Agricultural Sciences, 2019, 56(8): 1373-1381. |
| [13] | ZHU Xue-feng, TIAN Wen-gang, XIONG Xian-peng, XUE Fei, ZHANG Li, ZHU Hua-guo. Construction of CRISPR/Cas9 Expression Vector of GhPAO3 Gene in Cotton [J]. Xinjiang Agricultural Sciences, 2019, 56(2): 226-232. |
| [14] | Characteristics under Saline-alkali StressCHEN Kuan, MENG Chun-mei, SHI Jiao-hua, AN Meng-jie, MA Hong-xiu, WANG Kai-yong. Effects of Cotton Aphid on Cotton Growth and Physiological Characteristics under Saline-alkali Stress [J]. Xinjiang Agricultural Sciences, 2019, 56(11): 1988-1996. |
| [15] | WANG Long, LI Li-bei, MA Qi-feng, GENG Hong-wei, CHEN Quan-jia, QU Yan-ying, FAN Shu-li. Association Analysis of Verticillum Wilt Resistance Trait Using SSR Marker in Gossypium hirsutum L. [J]. Xinjiang Agricultural Sciences, 2018, 55(9): 1569-1578. |
| Viewed | ||||||||||||||||||||||||||||||||||||||||||||||||||
|
Full text 73
|
|
|||||||||||||||||||||||||||||||||||||||||||||||||
|
Abstract 247
|
|
|||||||||||||||||||||||||||||||||||||||||||||||||