

Xinjiang Agricultural Sciences ›› 2025, Vol. 62 ›› Issue (6): 1496-1506.DOI: 10.6048/j.issn.1001-4330.2025.06.022
• Soil Fertilizer·Plant Protection • Previous Articles Next Articles
LI Xinyan1( ), HUANG Tianyu2,3, WANG Zhiqiang2, ZHOU Hongxu1, WANG Bing2(
), HUANG Tianyu2,3, WANG Zhiqiang2, ZHOU Hongxu1, WANG Bing2( ), WANG Guirong2,4(
), WANG Guirong2,4( )
)
Received:2024-10-30
Online:2025-06-20
Published:2025-07-29
Correspondence author:
WANG Bing, WANG Guirong
Supported by:
李馨妍1( ), 黄天宇2,3, 王志强2, 周洪旭1, 王冰2(
), 黄天宇2,3, 王志强2, 周洪旭1, 王冰2( ), 王桂荣2,4(
), 王桂荣2,4( )
)
通讯作者:
王冰,王桂荣
作者简介:李馨妍(1999-),女,河北唐山人,硕士研究生,研究方向为化学生态学,(E-mail)lixinyan@stu.qau.edu.cn
基金资助:CLC Number:
LI Xinyan, HUANG Tianyu, WANG Zhiqiang, ZHOU Hongxu, WANG Bing, WANG Guirong. Identification and analysis of genes related to the synthesis and release of alarm pheromone of Megoura crassicauda[J]. Xinjiang Agricultural Sciences, 2025, 62(6): 1496-1506.
李馨妍, 黄天宇, 王志强, 周洪旭, 王冰, 王桂荣. 豌豆修尾蚜报警信息素合成与释放相关基因的鉴定与分析[J]. 新疆农业科学, 2025, 62(6): 1496-1506.
Add to citation manager EndNote|Ris|BibTeX
URL: https://www.xjnykx.com/EN/10.6048/j.issn.1001-4330.2025.06.022
| 基因ID Gene ID | 基因名称 Gene Name | FPKM-平均值 FPKM-Mean | 对数2倍数变化 log2FoldChange | P-值 P-value | 调整后P-值 P-adj | 上/下调 Up/Down- Regulation | |
|---|---|---|---|---|---|---|---|
| 残体 Residues | 腹管 Cornicles | ||||||
| MSTRG.2087.1 | McraCYP380C | 147.453 254 9 | 2902.503 026 | 4.299 216 63 | 3.22E-05 | 0.000 605 208 | Up |
| MSTRG.6860.1 | McraCYP4CK1 | 5 487.062 689 | 11 026.344 26 | 1.007 029 351 | 1.04E-17 | 2.15E-15 | Up |
| MSTRG.8070.6 | McraCYP315A1 | 5.033 902 502 | 518.314 676 4 | 6.685 247 751 | 2.80E-06 | 7.50E-05 | Up |
| MSTRG.11084.1 | McraCSP7 | 2 779.474 791 | 9 640.821 777 | 1.794 432 213 | 2.84E-156 | 6.04E-152 | Up |
| MSTRG.465.1 | McraCSP8 | 4 680.169 046 | 18 727.825 01 | 2.000 517 384 | 1.69E-34 | 1.57E-31 | Up |
| MSTRG.11160.2 | McraGPPS | 5 219 | 2 429 | -0.449 135 111 | 0.066 290 039 | 0.066 290 039 | - |
| MSTRG.7985.1 | McraFPPS | 8 660.666 667 | 3 161.666 667 | -0.597964673 | 1.48E-07 | 5.42E-06 | - |
Tab.1 Expression levels of candidate genes in the cornicles and residues of M. crassicauda
| 基因ID Gene ID | 基因名称 Gene Name | FPKM-平均值 FPKM-Mean | 对数2倍数变化 log2FoldChange | P-值 P-value | 调整后P-值 P-adj | 上/下调 Up/Down- Regulation | |
|---|---|---|---|---|---|---|---|
| 残体 Residues | 腹管 Cornicles | ||||||
| MSTRG.2087.1 | McraCYP380C | 147.453 254 9 | 2902.503 026 | 4.299 216 63 | 3.22E-05 | 0.000 605 208 | Up |
| MSTRG.6860.1 | McraCYP4CK1 | 5 487.062 689 | 11 026.344 26 | 1.007 029 351 | 1.04E-17 | 2.15E-15 | Up |
| MSTRG.8070.6 | McraCYP315A1 | 5.033 902 502 | 518.314 676 4 | 6.685 247 751 | 2.80E-06 | 7.50E-05 | Up |
| MSTRG.11084.1 | McraCSP7 | 2 779.474 791 | 9 640.821 777 | 1.794 432 213 | 2.84E-156 | 6.04E-152 | Up |
| MSTRG.465.1 | McraCSP8 | 4 680.169 046 | 18 727.825 01 | 2.000 517 384 | 1.69E-34 | 1.57E-31 | Up |
| MSTRG.11160.2 | McraGPPS | 5 219 | 2 429 | -0.449 135 111 | 0.066 290 039 | 0.066 290 039 | - |
| MSTRG.7985.1 | McraFPPS | 8 660.666 667 | 3 161.666 667 | -0.597964673 | 1.48E-07 | 5.42E-06 | - |
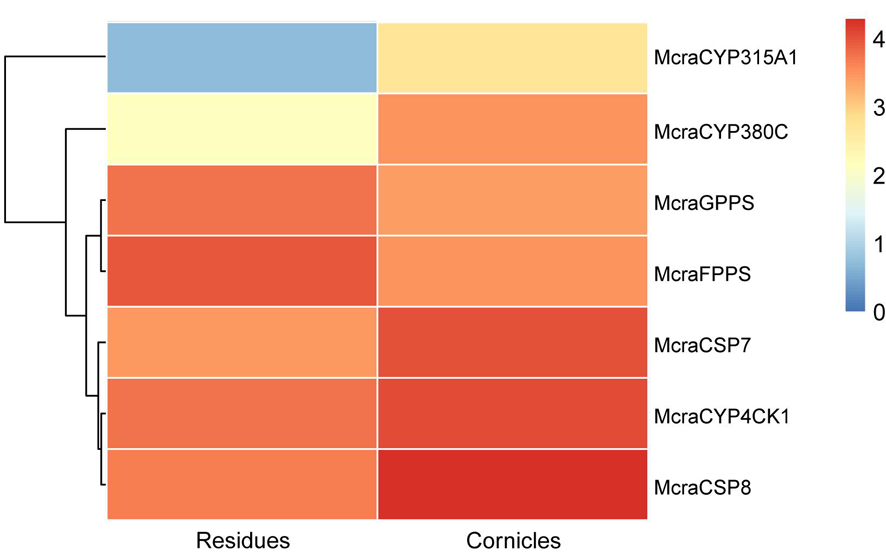
Fig.3 Expression profiles of candidate genes from M. crassicauda Notes: Mcra: M. crassicauda; Agos: A. gossypii; Apis: A. pisum; Pbam: P. bambucicola; The CYP4CK1 clade is highlighted with a red background, the CYP315A1 clade is marked with an orange background, and the CYP380C clade is indicated by a bluish purple background
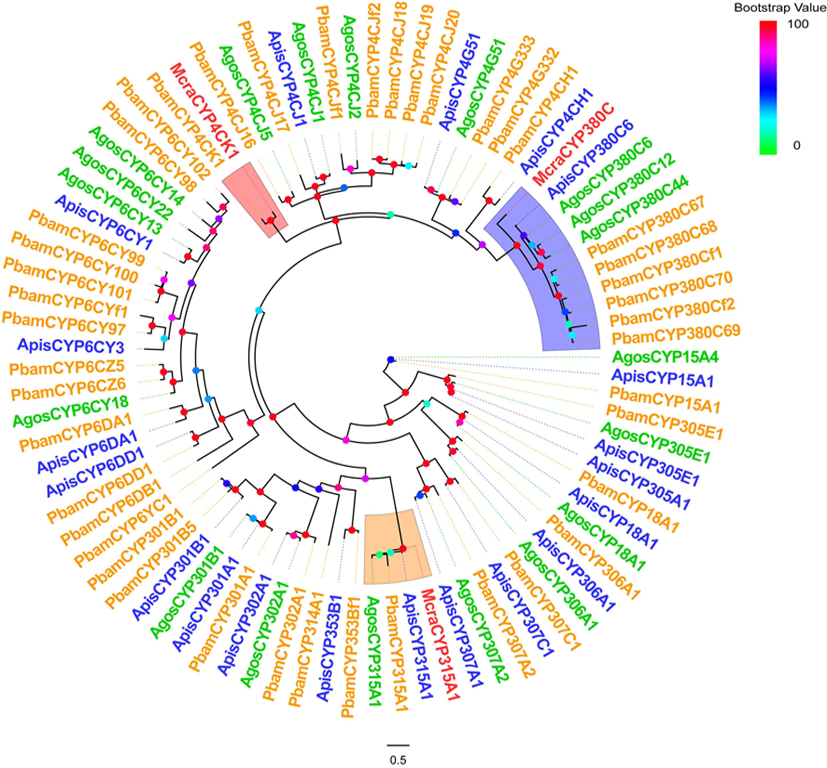
Fig.4 Phylogenetic tree of the CYP450s gene family of the four aphid species Notes: A. Amino acid sequence alignment between McraCYP380C and ApisCYP380C; B. Amino acid sequence alignment between McraCYP4CK1 and PbamCYP4CK1; C. Amino acid sequence alignment between McraCYP315A and ApisCYP315A1; Mcra: M. crassicauda; Apis: A. pisum; Pbam: P. bambucicola
| 参考基因 Unigene reference | 基因名 Gene name | 长度 Length (nt) | 开放 阅读框 ORF (aa) | E值 E- value | 相似度 Identity (%) | 最佳BLASTx匹配 Blastx best hit (Reference/Name/Species) | 信号肽 SP (No.) | 跨膜 结构域 TMD (No.) | 全长 Full length | 组织 Tissue |
|---|---|---|---|---|---|---|---|---|---|---|
| MSTRG.2087.1 | McraCYP380C | 1548 | 515 | 0.0 | 74.37 | >XP_060860450.1 PREDICTED: cytochrome P450 4C1-like isoform X1 [Metopolophium dirhodum] | 0 | 1 | YES | C&R |
| MSTRG.6860.1 | McraCYP4CK1 | 1547 | 515 | 0.0 | 89.57 | >XP_060860450.1 PREDICTED: cytochrome P450 4C1-like [Metopolophium dirhodum] | 0 | 1 | YES | C&R |
| MSTRG.8070.6 | McraCYP315A1 | 1434 | 477 | 0.0 | 90.78 | >XP_001944183.2 PREDICTED: cytochrome P450 315a1, mitochondrial [Acyrthosiphon pisum] | 0 | 0 | YES | C |
| MSTRG.11084.1 | McraCSP7 | 468 | 155 | 1e-109 | 100.00 | >ULF48248.1 PREDICTED: chemosensory protein 7 (CSP7) mRNA [Acyrthosiphon pisum] | 0 | 0 | YES | C&R |
| MSTRG.465.1 | McraCSP8 | 219 | 72 | 4e-27 | 100.00 | >ULF48249.1 PREDICTED: chemosensory protein 8 (CSP8) mRNA [Acyrthosiphon pisum] | - | - | 3’ | C&R |
| MSTRG.11160.2 | McraGPPS | 930 | 309 | 0.0 | 100.00 | >QUH22249.1 PREDICTED: geranyl pyrophosphate synthase mRNA [Megoura viciae] | 0 | 0 | YES | C&R |
| MSTRG.7985.1 | McraFPPS | 1185 | 394 | 0.0 | 98.98 | >AAY33489.2 PREDICTED: putative mitochondrial isoprenyl diphosphate synthase precursor, mRNA [Megoura viciae] | 0 | 0 | YES | C&R |
Tab.2 Annotated information of candidate genes
| 参考基因 Unigene reference | 基因名 Gene name | 长度 Length (nt) | 开放 阅读框 ORF (aa) | E值 E- value | 相似度 Identity (%) | 最佳BLASTx匹配 Blastx best hit (Reference/Name/Species) | 信号肽 SP (No.) | 跨膜 结构域 TMD (No.) | 全长 Full length | 组织 Tissue |
|---|---|---|---|---|---|---|---|---|---|---|
| MSTRG.2087.1 | McraCYP380C | 1548 | 515 | 0.0 | 74.37 | >XP_060860450.1 PREDICTED: cytochrome P450 4C1-like isoform X1 [Metopolophium dirhodum] | 0 | 1 | YES | C&R |
| MSTRG.6860.1 | McraCYP4CK1 | 1547 | 515 | 0.0 | 89.57 | >XP_060860450.1 PREDICTED: cytochrome P450 4C1-like [Metopolophium dirhodum] | 0 | 1 | YES | C&R |
| MSTRG.8070.6 | McraCYP315A1 | 1434 | 477 | 0.0 | 90.78 | >XP_001944183.2 PREDICTED: cytochrome P450 315a1, mitochondrial [Acyrthosiphon pisum] | 0 | 0 | YES | C |
| MSTRG.11084.1 | McraCSP7 | 468 | 155 | 1e-109 | 100.00 | >ULF48248.1 PREDICTED: chemosensory protein 7 (CSP7) mRNA [Acyrthosiphon pisum] | 0 | 0 | YES | C&R |
| MSTRG.465.1 | McraCSP8 | 219 | 72 | 4e-27 | 100.00 | >ULF48249.1 PREDICTED: chemosensory protein 8 (CSP8) mRNA [Acyrthosiphon pisum] | - | - | 3’ | C&R |
| MSTRG.11160.2 | McraGPPS | 930 | 309 | 0.0 | 100.00 | >QUH22249.1 PREDICTED: geranyl pyrophosphate synthase mRNA [Megoura viciae] | 0 | 0 | YES | C&R |
| MSTRG.7985.1 | McraFPPS | 1185 | 394 | 0.0 | 98.98 | >AAY33489.2 PREDICTED: putative mitochondrial isoprenyl diphosphate synthase precursor, mRNA [Megoura viciae] | 0 | 0 | YES | C&R |

Fig.5 Amino acid sequence alignment of McraCYP450s genes from M. crassicauda with homologous genes of other aphid species Notes: Mcra: M. crassicauda; Agos: A. gossypii; Apis: A. pisum; Agly: A. glycines; Mper: M. persicae; Save: S. avenae; The candidate CSP genes identified from the cornicles of M. crassicauda are indicated by black boxes. The CSP7 clade is highlighted with a light red background, while the CSP8 clade is high-lighted with a purple background
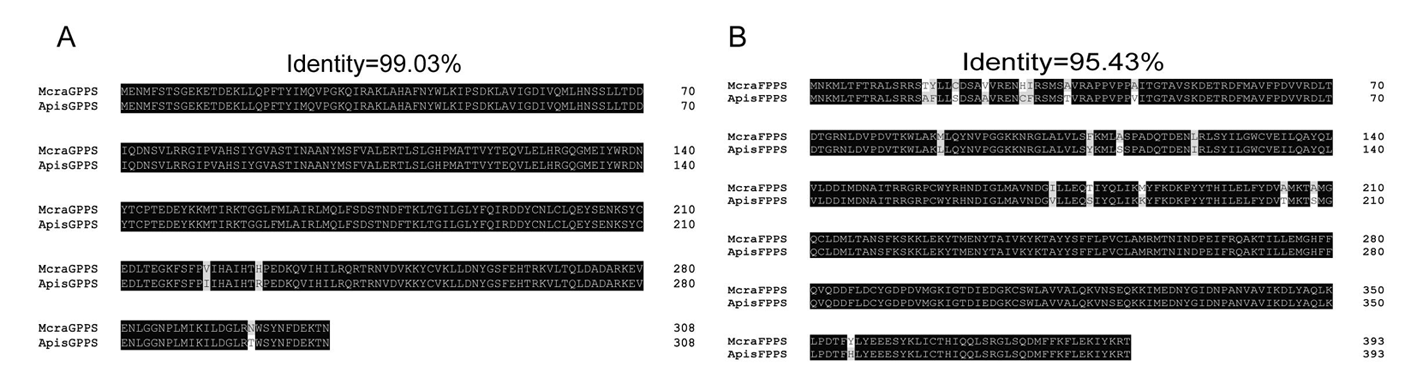
Fig.7 Amino acid sequence alignment of McraIDS genes from M. crassicauda with homologous genes of A. pisum Notes: A. Sequence alignment between McraGPPS and ApisGPPS; B. Sequence alignment between McraFPPS and ApisFPPS; Mcra: M. crassicauda; Apis: A. pisum
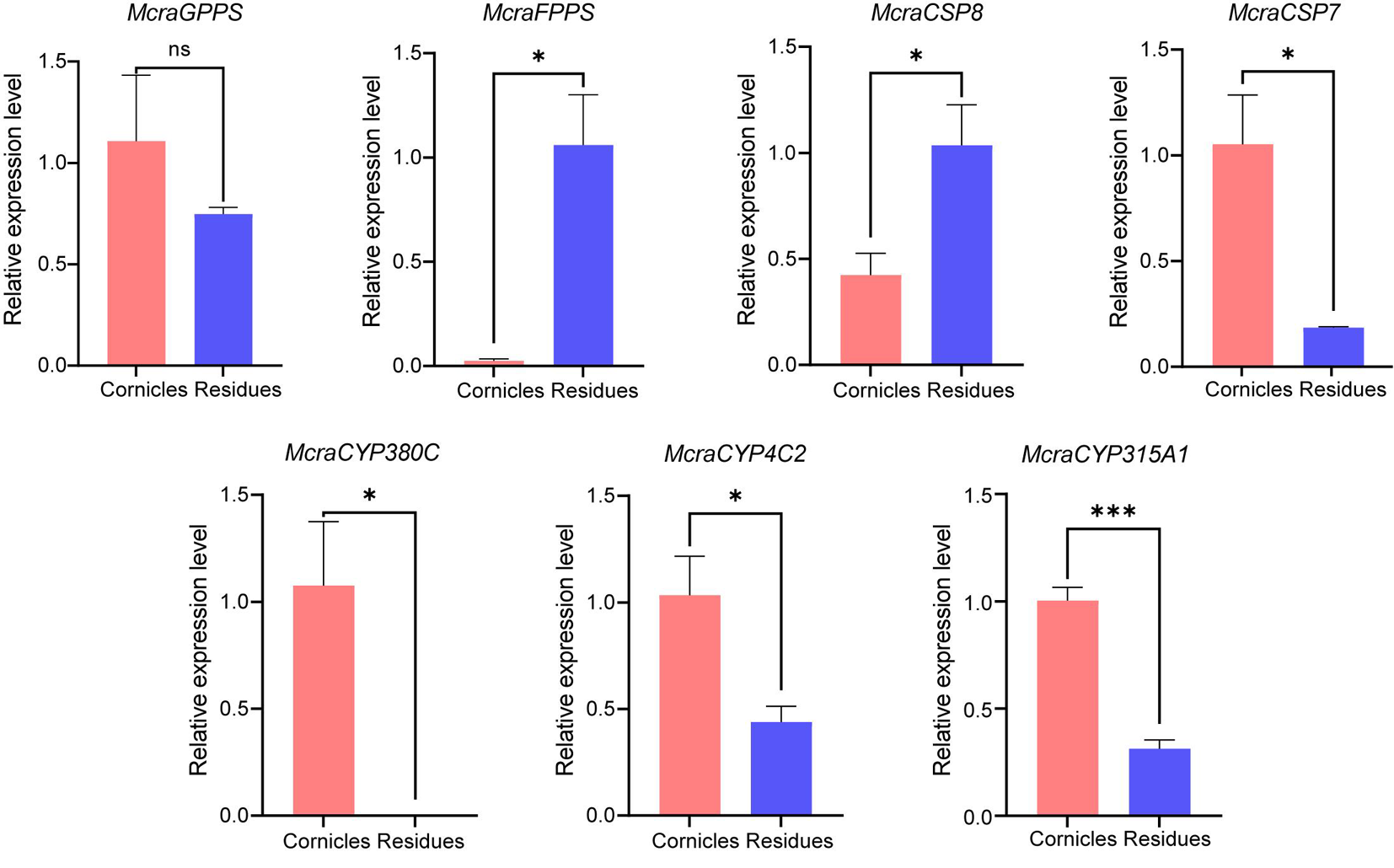
Fig.8 T Tissue-specific expression levels of seven candidate genes from M. crassicauda Notes: Data are presented as Mean±SE (n=3). The relative expression between cornicles and residues was analyzed by t-test, asterisks indicate a significant difference at the α=0.05 level, while n.s. indicates no significant difference at the α=0.05 level
| [1] | Weisser W W, Braendle C, Minoretti N. Predator-induced morphological shift in the pea aphid[J]. Proceedings of the Royal Society of London Series B: Biological Sciences, 1999, 266(1424): 1175-1181. |
| [2] | Badji C A, Sol-Mochkovitch Z, Fallais C, et al. Alarm pheromone responses depend on genotype, but not on the presence of facultative endosymbionts in the pea aphid Acyrthosiphon pisum[J]. Insects, 2021, 12(1): 43. |
| [3] | Chen S W, Edwards J S. Observations on the structure of secretory cells associated with aphid cornicles[J]. Zeitschrift Fur Zellforschung und Mikroskopische Anatomie, 1972, 130(3): 312-317. |
| [4] | Vandermoten S, Mescher M C, Francis F, et al. Aphid alarm pheromone: an overview of current knowledge on biosynthesis and functions[J]. Insect Biochemistry and Molecular Biology, 2012, 42(3): 155-163. |
| [5] | Micha S G, Wyss U. Aphid alarm pheromone (E)-β-farnesene: a host finding kairomone for the aphid primary parasitoidAphidius uzbekistanicus (Hymenoptera: Aphidiinae)[J]. CHEMOECOLOGY, 1996, 7(3): 132-139. |
| [6] | Bowers W S, Nault L R, Webb R E, et al. Aphid alarm pheromone: isolation, identification, synthesis[J]. Science, 1972, 177(4054): 1121-1122. |
| [7] | Pickett J A, Wadhams L J, Woodcock C M, et al. The chemical ecology of aphids[J]. Annual Review of Entomology, 1992, 37: 67-90. |
| [8] | Edwards L J, Siddall J B, Dunham L L, et al. Trans-β-farnesene, alarm pheromone of the green peach aphid, Myzus persicae (Sulzer)[J]. Nature, 1973, 241(5385): 126-127. |
| [9] | Pickett J A, Griffiths D C. Composition of aphid alarm pheromones[J]. Journal of Chemical Ecology, 1980, 6(2): 349-360. |
| [10] | Wang B, Dong W Y, Li H M, et al. Molecular basis of (E)-β-farnesene-mediated aphid location in the predator Eupeodes corollae[J]. Current Biology, 2022, 32(5): 951-962.e7. |
| [11] | Lewis M J, Prosser I M, Mohib A, et al. Cloning and characterisation of a prenyltransferase from the aphid Myzus persicae with potential involvement in alarm pheromone biosynthesis[J]. Insect Molecular Biology, 2008, 17(4): 437-443. |
| [12] | Sun Z J, Li Z X. The terpenoid backbone biosynthesis pathway directly affects the biosynthesis of alarm pheromone in the aphid[J]. Insect Molecular Biology, 2018, 27(6): 824-834. |
| [13] | Ma G Y, Sun X F, Zhang Y L, et al. Molecular cloning and characterization of a prenyltransferase from the cotton aphid, Aphis gossypii[J]. Insect Biochemistry and Molecular Biology, 2010, 40(7): 552-561. |
| [14] | Sun X F, Li Z X. In silico and in vitro analyses identified three amino acid residues critical to the catalysis of two aphid farnesyl diphosphate synthase[J]. The Protein Journal, 2012, 31(5): 417-424. |
| [15] | Song X, Qin Y G, Zhang Y H, et al. Farnesyl/geranylgeranyl diphosphate synthases regulate the biosynthesis of alarm pheromone in a unique manner in the vetch aphid Megoura viciae[J]. Insect Molecular Biology, 2023, 32(3): 229-239. |
| [16] | Beran F, Rahfeld P, Luck K, et al. Novel family of terpene synthases evolved from trans-isoprenyl diphosphate synthases in a flea beetle[J]. Proceedings of the National Academy of Sciences of the United States of America, 2016, 113(11): 2922-2927. |
| [17] | Lancaster J, Khrimian A, Young S, et al. De novo formation of an aggregation pheromone precursor by an isoprenyl diphosphate synthase-related terpene synthase in the harlequin bug[J]. Proceedings of the National Academy of Sciences of the United States of America, 2018, 115(37): E8634-E8641. |
| [18] | Lu K, Song Y Y, Zeng R S. The role of cytochrome P450-mediated detoxification in insect adaptation to xenobiotics[J]. Current Opinion in Insect Science, 2021, 43: 103-107. |
| [19] | Sezutsu H, Le Goff G, Feyereisen R. Origins of P450 diversity[J]. Philosophical Transactions of the Royal Society B: Biological Sciences, 2013, 368(1612): 20120428. |
| [20] | Song M M, Kim A C, Gorzalski A J, et al. Functional characterization of myrcene hydroxylases from two geographically distinct Ips pini populations[J]. Insect Biochemistry and Molecular Biology, 2013, 43(4): 336-343. |
| [21] | Fu N X, Yang Z L, Pauchet Y, et al. A cytochrome P450 from the mustard leaf beetles hydroxylates geraniol, a key step in iridoid biosynthesis[J]. Insect Biochemistry and Molecular Biology, 2019, 113: 103212. |
| [22] | Wang Q, Zhou J J, Liu J T, et al. Integrative transcriptomic and genomic analysis of odorant binding proteins and chemosensory proteins in aphids[J]. Insect Molecular Biology, 2019, 28(1): 1-22. |
| [23] | Wenger J A, Cassone B J, Legeai F, et al. Whole genome sequence of the soybean aphid, Aphis glycines[J]. Insect Biochemistry and Molecular Biology, 2020, 123: 102917. |
| [24] | Lu J J, Zhang H, Wang Q, et al. Genome-wide identification and expression pattern of cytochrome P450 genes in the social aphid Pseudoregma bambucicola[J]. Insects, 2023, 14(2): 212. |
| [25] | Gauthier J P, Legeai F, Zasadzinski A, et al. AphidBase: a database for aphid genomic resources[J]. Bioinformatics, 2007, 23(6): 783-784. |
| [26] | Kislow C J, Edwards L J. Repellent Odour in Aphids[J]. Nature, Nature Publishing Group, 1972, 235(5333): 108-109. |
| [27] | Gut J, van Oosten A M. Functional significance of the alarm pheromone composition in various morphs of the green peach aphid, Myzus persicae[J]. Entomologia Experimentalis et Applicata, 1985, 37(2): 199-204. |
| [28] | Grabherr M G, Haas B J, Yassour M, et al. Full-length transcriptome assembly from RNA-Seq data without a reference genome[J]. Nature Biotechnology, 2011, 29(7): 644-652. |
| [29] | Pertea G, Huang X Q, Liang F, et al. TIGR Gene Indices clustering tools (TGICL): a softwaresystem for fast clustering of large EST datasets[J]. Bioinformatics, 2003, 19(5): 651-652. |
| [30] | Wang B, Liu Y, Wang G R. Chemosensory genes in the antennal transcriptome of two syrphid species, Episyrphus balteatus and Eupeodes corollae (Diptera: Syrphidae)[J]. BMC Genomics, 2017, 18(1): 586. |
| [31] | Liu Y P, Cui Z Y, Si P F, et al. Characterization of a specific odorant receptor for linalool in the Chinese Citrus fly Bactrocera minax (Diptera: Tephritidae)[J]. Insect Biochemistry and Molecular Biology, 2020, 122: 103389. |
| [32] | Conesa A, Götz S, García-Gómez J M, et al. Blast2GO: a universal tool for annotation, visualization and analysis in functional genomics research[J]. Bioinformatics, 2005, 21(18): 3674-3676. |
| [33] | Gasteiger E, Gattiker A, Hoogland C, et al. ExPASy: The proteomics server for in-depth protein knowledge and analysis[J]. Nucleic Acids Research, 2003, 31(13): 3784-3788. |
| [34] | Petersen T N, Brunak S, von Heijne G, et al. SignalP 4.0: discriminating signal peptides from transmembrane regions[J]. Nature Methods, 2011, 8(10): 785-786. |
| [35] | Krogh A, Larsson B, von Heijne G, et al. Predicting transmembrane protein topology with a hidden Markov model: application to complete genomes[J]. Journal of Molecular Biology, 2001, 305(3): 567-580. |
| [36] | Katoh K, Standley D M. MAFFT multiple sequence alignment software version 7: improvements in performance and usability[J]. Molecular Biology and Evolution, 2013, 30(4): 772-780. |
| [37] | Stamatakis A. RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies[J]. Bioinformatics, 2014, 30(9): 1312-1313. |
| [38] | Wang B, Huang T Y, Yao Y, et al. A conserved odorant receptor identified from antennal transcriptome of Megoura crassicauda that specifically responds to Cis-jasmone[J]. Journal of Integrative Agriculture, 2022, 21(7): 2042-2054. |
| [39] | Li B, Dewey C N. RSEM: accurate transcript quantification from RNA-Seq data with or without a reference genome[J]. BMC Bioinformatics, 2011, 12: 323. |
| [40] | Wickham H. ggplot2: Elegant Graphics for Data Analysis[M]. Springer Publishing Company, Incorporated, 2009. |
| [41] | Ishikawa A, Ishikawa Y, Okada Y, et al. Screening of upregulated genes induced by high density in the vetch aphid Megoura crassicauda[J]. Journal of Experimental Zoology Part A, Ecological Genetics and Physiology, 2012, 317(3): 194-203. |
| [42] | Pfaffl M W. A new mathematical model for relative quantification in real-time RT-PCR[J]. Nucleic Acids Research, 2001, 29(9): e45. |
| [43] | Livak K J, Schmittgen T D. Analysis of relative gene expression data using real-time quantitative PCR and the 2 (-Delta Delta C(T)) Method[J]. Methods, 2001, 25(4): 402-408. |
| [44] | Angeli S, Ceron F, Scaloni A, et al. Purification, structural characterization, cloning and immunocytochemical localization of chemoreception proteins from Schistocerca gregaria[J]. European Journal of Biochemistry, 1999, 262(3): 745-754. |
| [45] | Butler P J, Harris J I, Hartley B S, et al. Reversible blocking of peptide amino groups by maleic anhydride[J]. The Biochemical Journal, 1967, 103(3): 78P-79P. |
| [46] | Waschkuhn A. Robert A. Dahl, Polyarchy: Participation and Opposition, New Haven 1971[A]. In: S. Kailitz. Schlüsselwerke der Politikwissenschaft[M]. Wiesbaden: VS Verlag für Sozialwissenschaften, 2007: 86-88. |
| [47] | Rane R V, Ghodke A B, Hoffmann A A, et al. Detoxifying enzyme complements and host use phenotypes in 160 insect species[J]. Current Opinion in Insect Science, 2019, 31: 131-138. |
| [48] | Sun C X, Li Z X. Biosynthesis of aphid alarm pheromone is modulated in response to starvation stress under regulation by the insulin, glycolysis and isoprenoid pathways[J]. Journal of Insect Physiology, 2021, 128: 104174. |
| [49] | Zhang H, Li Z X. A type-III insect geranylgeranyl diphosphate synthase with a novel catalytic property[J]. Protein and Peptide Letters, 2014, 21(7): 615-623. |
| [50] | Cheng Y J, Li Z X. Spatiotemporal expression profiling of the farnesyl diphosphate synthase genes in aphids and analysis of their associations with the biosynthesis of alarm pheromone[J]. Bulletin of Entomological Research, 2019, 109(3): 398-407. |
| [51] | Truman J W, Riddiford L M. Endocrine insights into the evolution of metamorphosis in insects[J]. Annual Review of Entomology, 2002, 47: 467-500. |
| [52] | Picimbon J F, Dietrich K, Krieger J, et al. Identity and expression pattern of chemosensory proteins in Heliothis virescens (Lepidoptera, Noctuidae)[J]. Insect Biochemistry and Molecular Biology, 2001, 31(12): 1173-1181. |
| [53] | Pelosi P, Iovinella I, Zhu J, et al. Beyond chemoreception: diverse tasks of soluble olfactory proteins in insects[J]. Biological Reviews of the Cambridge Philosophical Society, 2018, 93(1): 184-200. |
| [54] | Nomura Kitabayashi A, Arai T, Kubo T, et al. Molecular cloning of cDNA for p10, a novel protein that increases in the regenerating legs of Periplaneta americana (American cockroach)[J]. Insect Biochemistry and Molecular Biology, 1998, 28(10): 785-790. |
| [1] | WANG Yaling, JIANG Yinghong, SUN Hui, LIU Yi. Comparative transcriptome analysis between two potato varieties with different salt-tolerance and further identification of potato salt-tolerance genes [J]. Xinjiang Agricultural Sciences, 2025, 62(5): 1121-1130. |
| [2] | JU Le, QI Juncang, NIU Yinting, SHI Peichun, SONG Ruijiao, SONG Lingyu, YIN Zhigang, CHEN Peiyu, QIANG Xuelan. RNA-seq-based mining and analysis of drought-related genes in barley seedlings [J]. Xinjiang Agricultural Sciences, 2024, 61(5): 1077-1084. |
| [3] | HE Wanjie, MENG Hanying, ZHI Mengting, CHEN Jing. Analysis of the antennal transcriptome and differentially expressed genes of female and male Monolepta signata [J]. Xinjiang Agricultural Sciences, 2024, 61(4): 984-995. |
| [4] | HU Jinge, BAI Shijian, CHEN Guang, CAI Junshe. Transcriptome and metabolome integrated analysis of flavonoids in Xinyu grape peel under different ground mulch 5ypes [J]. Xinjiang Agricultural Sciences, 2024, 61(1): 63-78. |
| [5] | WEI Wei, SHAN Shouming, XU Wendi, LI Guangzong. Transcriptome analysis of callus at rooting stage in tissue culture of vitis amurensis 'shuangyou' [J]. Xinjiang Agricultural Sciences, 2023, 60(6): 1451-1459. |
| [6] | SONG Jindi, LIU Jun, SUN Yufang, Youlituzi Naibi, CHEN Baoqiang, XIE Bingbing. Transcriptome Analysis of Acidovorax citrulli against Copper Stress [J]. Xinjiang Agricultural Sciences, 2023, 60(2): 389-398. |
| [7] | WANG Yejian, YANG Jie, LIANG Xiaoling, Abulati Abra, HAN Denxu, XI Haojiang, LIU Jun, LI Mingdong. Transcriptome Analysis of Floral Organ Differentiation Stage of Two Maize Inbred Lines under Drought Stress [J]. Xinjiang Agricultural Sciences, 2020, 57(9): 1578-1585. |
| [8] | WANG Zhen-Dong, LU Xiao-Yan, TU Wen-Wen, HE Chen-Chen. Transcriptome Sequencing Analysis of Leaf and Root of Sour Jujube Seedlings under NaCl Stress Alleviated by Exogenous CaCl2 [J]. Xinjiang Agricultural Sciences, 2019, 56(6): 1052-1062. |
| [9] | ZHANG Yi-yuan, GUO Yang-hua, WANG Cong-hui, TANG Hong, NAN Hai-yan, WANG Li-min, ZHOU Pin. Induction and Transcriptomics Analysis of Ovine iPS Cells [J]. Xinjiang Agricultural Sciences, 2018, 55(11): 2142-2149. |
| [10] | BAO Qiu-juan, ZHANG Li-li, Hainar Wulazibai, ZHANG Fu-chun. Analysis of DNA Damage Repair Related Genes in Drought Stress Cotton Transcriptome [J]. Xinjiang Agricultural Sciences, 2017, 54(11): 1999-2005. |
| Viewed | ||||||
|
Full text |
|
|||||
|
Abstract |
|
|||||