

新疆农业科学 ›› 2022, Vol. 59 ›› Issue (6): 1312-1330.DOI: 10.6048/j.issn.1001-4330.2022.06.002
宋吉坤1,2( ), 辛玥1, 李龙云1, 刘国元1, 裴文锋1,2, 马建江1, 曲延英2, 于霁雯1,2(
), 辛玥1, 李龙云1, 刘国元1, 裴文锋1,2, 马建江1, 曲延英2, 于霁雯1,2( ), 吴嫚1(
), 吴嫚1( )
)
收稿日期:2021-09-26
出版日期:2022-06-20
发布日期:2022-07-07
通信作者:
于霁雯(1978-),女,河南安阳人,研究员,博士,研究方向为棉花分子育种,(E-mail) yujw666@hotmail.com;作者简介:宋吉坤(1995-),男,山东青岛人,博士研究生,研究方向为棉花分子育种,(E-mail) sjkow513@163.com
基金资助:
SONG Jikun1,2( ), XIN Yue1, LI Longyun1, LIU Guoyuan1, PEI Wenfeng1,2, MA Jianjiang1, QU Yanying2, YU Jiwen1,2(
), XIN Yue1, LI Longyun1, LIU Guoyuan1, PEI Wenfeng1,2, MA Jianjiang1, QU Yanying2, YU Jiwen1,2( ), WU Man1(
), WU Man1( )
)
Received:2021-09-26
Published:2022-06-20
Online:2022-07-07
Supported by:摘要:
【目的】 利用适应性广、产量高的陆地棉和纤维品质优异的海岛棉进行高通量基因芯片比较分析,筛选和鉴定棉纤维发育相关的关键基因。【方法】 以SG747(Gossypium hirsutum L.)和Giza75(Gossypium barbadense L.)为材料,利用Affymetrix公司的棉花寡聚核苷酸基因芯片对其开花后10 d(10 days after anthesis, 10 DPA)的棉纤维进行表达谱分析。【结果】 材料Giza75和SG747共筛选到3 905个差异表达基因,其中功能预测基因占比17.80%,翻译、核糖体结构相关基因占比16.82%,翻译后修饰占比14.32%。Gra.2198.1.A1_at正向调控棉纤维发育过程,Gra.85.1.S1_at、Ghi.249.1.A1_at和Ghi.8448.1.S1_x_at负向调控棉纤维发育过程,可能参与了棉纤维发育过程。【结论】 利用陆地棉和海岛棉基因芯片并结合qRT-PCR筛选和鉴定到4个纤维发育相关的关键候选基因。
中图分类号:
宋吉坤, 辛玥, 李龙云, 刘国元, 裴文锋, 马建江, 曲延英, 于霁雯, 吴嫚. 利用基因芯片分析比较Giza75和SG747纤维发育差异表达基因[J]. 新疆农业科学, 2022, 59(6): 1312-1330.
SONG Jikun, XIN Yue, LI Longyun, LIU Guoyuan, PEI Wenfeng, MA Jianjiang, QU Yanying, YU Jiwen, WU Man. Comparative Analysis of Gene Expression between Giza 75 and SG 747 Using Cotton Oligonucleotide Microarrays during Fiber Development[J]. Xinjiang Agricultural Sciences, 2022, 59(6): 1312-1330.
| 探针编号 Probe Set | NCBI 登录号 Accessionnumber | 基因名称 Gene Name | 引物序列 Sequence | 引物长度 Length (bp) |
|---|---|---|---|---|
| __ | AY305737 | Actin9 | -F5’-ATCCTCCGTCTTGACCTTG -3’ | 19 |
| -R5’-TGTCCGTCAGGCAACTCAT-3’ | 19 | |||
| Ghi.8448.1.S1_x_at | AF521240.1 | GhXu-142 βTUB1 | -F5’-TAACTATGTCGGCACTTC -3’ | 18 |
| -R5’-AATCAATTCAGCTCCTTC -3’ | 18 | |||
| Gra.2198.1.A1_at | CO100609 | GhSAHH | -F5’-CAGCTCCCAGATCCGTCTTC -3’ | 20 |
| -R5’-CACCAACTAATCTCTCCCTCATCC -3’ | 24 | |||
| Gra.85.1.S1_at | CO097359 | GhCESA4 | -F5’- GTAACCATAAGGCATGTCCAAAATG-3’ | 25 |
| -R5’- AGAAAAGGTGGGAATGAACGAA-3’ | 22 | |||
| Ghi.249.1.A1_at | DQ204496.1 | GhAlpha-expansin 2 | -F5’-CCCCTGTATGAAGAAAGGAGGA -3’ | 22 |
| -R5’-AACATCTCCAGCACCACCAAC -3’ | 21 | |||
| -R5’-CGAAGTTGTTGGCAGCGTCT -3’ | 20 |
表1 qRT-PCR所用基因及其引物序列
Table 1 Gene Primers for quantitative RT-PCR analysis
| 探针编号 Probe Set | NCBI 登录号 Accessionnumber | 基因名称 Gene Name | 引物序列 Sequence | 引物长度 Length (bp) |
|---|---|---|---|---|
| __ | AY305737 | Actin9 | -F5’-ATCCTCCGTCTTGACCTTG -3’ | 19 |
| -R5’-TGTCCGTCAGGCAACTCAT-3’ | 19 | |||
| Ghi.8448.1.S1_x_at | AF521240.1 | GhXu-142 βTUB1 | -F5’-TAACTATGTCGGCACTTC -3’ | 18 |
| -R5’-AATCAATTCAGCTCCTTC -3’ | 18 | |||
| Gra.2198.1.A1_at | CO100609 | GhSAHH | -F5’-CAGCTCCCAGATCCGTCTTC -3’ | 20 |
| -R5’-CACCAACTAATCTCTCCCTCATCC -3’ | 24 | |||
| Gra.85.1.S1_at | CO097359 | GhCESA4 | -F5’- GTAACCATAAGGCATGTCCAAAATG-3’ | 25 |
| -R5’- AGAAAAGGTGGGAATGAACGAA-3’ | 22 | |||
| Ghi.249.1.A1_at | DQ204496.1 | GhAlpha-expansin 2 | -F5’-CCCCTGTATGAAGAAAGGAGGA -3’ | 22 |
| -R5’-AACATCTCCAGCACCACCAAC -3’ | 21 | |||
| -R5’-CGAAGTTGTTGGCAGCGTCT -3’ | 20 |
| 性状 Trait | 基因型Genetype | T测验T test | |||
|---|---|---|---|---|---|
| Giza75 | SG7474 | T Stat | P(T≤T)双尾 P(T≤t) two- tailed | ||
| 纤维长度 Fiber Length (mm) | 34.090 0 | 29.201 7 | 10.433 0 | 0.000 0 | ** |
| 整齐度指数 Uniformity index(%) | 86.066 7 | 84.666 7 | 2.613 8 | 0.028 1 | * |
| 马克隆值 Micronair value | 4.565 0 | 5.621 7 | -3.038 0 | 0.028 8 | * |
| 伸长率 Fiber Elongation rate(%) | 5.750 0 | 6.416 7 | -1.913 0 | 0.092 1 | |
| 纤维强度 Fiber Strength (cN/tex) | 40.391 3 | 27.693 3 | 8.487 3 | 0.000 0 | ** |
| 籽棉产量 Seed Yield (kg/667m2) | 70.947 5 | 191.813 8 | -4.838 0 | 0.000 7 | ** |
| 皮棉产量 Lint Yield (kg/667m2) | 26.677 5 | 78.272 5 | -5.397 7 | 0.000 2 | ** |
| 衣分 Lint Percentage (%) | 36.926 3 | 41.707 5 | -4.312 8 | 0.000 8 | ** |
| 铃重 Boll Weight (g) | 3.617 5 | 6.007 5 | -10.792 4 | 0.000 0 | ** |
表2 陆地棉与海岛棉的纤维品质和产量性状差异
Table 2 Fiber quality andyield difference between G. hirsutum and G. barbadense
| 性状 Trait | 基因型Genetype | T测验T test | |||
|---|---|---|---|---|---|
| Giza75 | SG7474 | T Stat | P(T≤T)双尾 P(T≤t) two- tailed | ||
| 纤维长度 Fiber Length (mm) | 34.090 0 | 29.201 7 | 10.433 0 | 0.000 0 | ** |
| 整齐度指数 Uniformity index(%) | 86.066 7 | 84.666 7 | 2.613 8 | 0.028 1 | * |
| 马克隆值 Micronair value | 4.565 0 | 5.621 7 | -3.038 0 | 0.028 8 | * |
| 伸长率 Fiber Elongation rate(%) | 5.750 0 | 6.416 7 | -1.913 0 | 0.092 1 | |
| 纤维强度 Fiber Strength (cN/tex) | 40.391 3 | 27.693 3 | 8.487 3 | 0.000 0 | ** |
| 籽棉产量 Seed Yield (kg/667m2) | 70.947 5 | 191.813 8 | -4.838 0 | 0.000 7 | ** |
| 皮棉产量 Lint Yield (kg/667m2) | 26.677 5 | 78.272 5 | -5.397 7 | 0.000 2 | ** |
| 衣分 Lint Percentage (%) | 36.926 3 | 41.707 5 | -4.312 8 | 0.000 8 | ** |
| 铃重 Boll Weight (g) | 3.617 5 | 6.007 5 | -10.792 4 | 0.000 0 | ** |
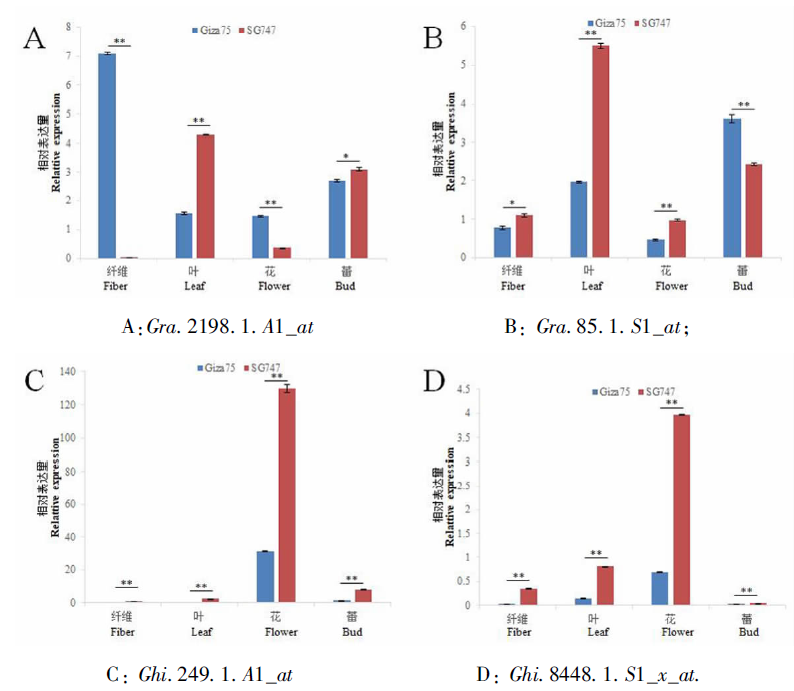
图3 差异表达基因在不同组织中的qRT-PCR 注:纤维为开花后5 d的纤维;*和 **分别表示在0.05和0.01显著水平差异
Fig.3 qRT-PCR analysis results of differentially expressed genes in different tissues. Note: The fiber is the fiber at 5 days after flowering; * and ** indicate significant differences at 0.05 and 0.01 levels, respectively.
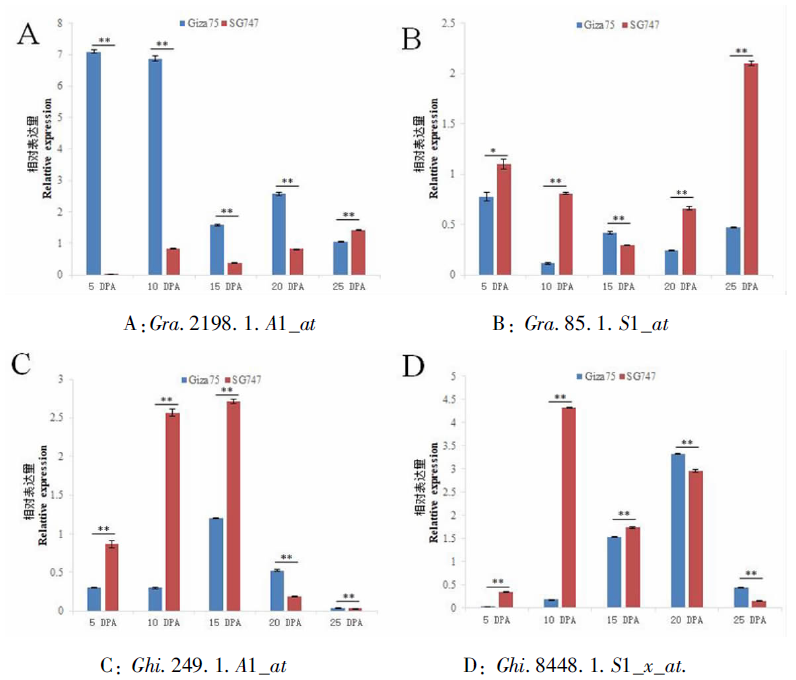
图4 差异表达基因在纤维不同发育时期的qRT-PCR 注:DPA为开花后的天数;*和**分别表示在0.05和0.01显著水平差异
Fig.4 qRT-PCR analysis results of differentially expressed genes at different developmental stages of fiber Note: DPA is the number of days after flowering; * and ** indicate significant differences at 0.05 and 0.01 levels, respectively
| [1] | Wendel J F, Brubaker C, Alvarez I, et al. Evolution and natural history of the cotton genus[J]. In: Paterson AP (ed.) Genetics and Genomics of Cotton, 2009, 3-22. |
| [2] | Bowman D T, Gutierrez O A. Sources of fiber strength in the U.S. upland cotton crop from 1980 to 2000[J]. The Journal of Cotton Science, 2003, 7:164-169. |
| [3] |
Sajid M, Ahmad R I, Muhammad A R, et al. Role of SNPs in determining QTLs for major traits in cotton[J]. Journal of Cotton Research, 2019, 2:5.
DOI URL |
| [4] |
Ma L L, Su Y, Nie H S, et al. QTL and genetic analysis controlling fiber quality traits using paternal back cross population in upland cotton[J]. Journal of Cotton Research, 2020, 3:22.
DOI URL |
| [5] |
Deng X Y, Gong J W, Liu A Y, et al. QTL mapping for fiber quality and yield related traits across multiple generations in segregating population of CCRI 70[J]. Journal of Cotton Research, 2019, 2:13.
DOI URL |
| [6] |
Tu L L, Zhang X L, Liang S G, et al. Genes expression analyses of sea-island cotton (Gossypium barbadense L.) during fiber development[J]. Plant Cell Reports, 2007, 26(8):1309-1320.
PMID |
| [7] |
Wu M, Zhang L Y, Li X L, et al. A comparative transciptome analysis of two sets of backcross inbred lines differing in lint-yield derived from a Gossypium hirsutum × Gossypium barbadense population[J]. Molecular Genetics and Genomics, 2016, 291(4):1749-1767.
DOI URL |
| [8] |
Alabady M S, Youn E, Wilkins T A. Double feature selection and cluster analyses in mining of microarray data from cotton[J]. BMC Genomics, 2008, 9:295.
DOI PMID |
| [9] |
Wu M, Li L Y, Liu G Y, et al. Differentially expressed genes between two groups of backcross inbred lines differing in fiber length developed from Upland × Pima cotton[J]. Molecular Biology Reports, 2019, 46(1):1199-1212.
DOI URL |
| [10] |
Qin Y M, Zhu Y X. How cotton fibers elongate: a tale of linear cell-growth mode[J]. Current Opinion in Plant Biology, 2011, 14(1):106-11.
DOI URL |
| [11] |
Liu C X, Li Z F, Dou L L, et al. A genome-wide identification of the BLH gene family reveals BLH1 involved in cotton fiber development[J]. Journal of Cotton Research, 2020, 3:26.
DOI URL |
| [12] |
Chen F, Guo Y J, Chen L, et al. Global identification of genes associated with xylan biosynthesis in cotton fiber[J]. Journal of Cotton Research, 2020, 3:25.
DOI URL |
| [13] |
Yu J W, Zhang K, Li S Y, et al. Mapping quantitative trait loci for lint yield and fiber quality across environments in a Gossypium hirsutum × Gossypium barbadense backcross inbred line population[J]. Theoretical and Applied Genetics, 2013, 126(1): 275-287.
DOI URL |
| [14] | 蒋建雄, 张天真. 利用CTAB/酸酚法提取棉花组织总RNA[J]. 棉花学报, 2003, 15(3):166-167. |
| JIANG Jiangxiong, ZHANG Tianzhen. Extraction of total RNA in cotton tissues with CTAB-acidic phenolic method[J]. Cotton Science, 2003, 15(3):166-167 | |
| [15] | 李龙云, 于霁雯, 翟红红, 等. 利用基因芯片技术筛选棉纤维伸长相关基因[J]. 作物学报, 2011, 37(1):95-104. |
|
LI Longyun, YU Jiwen, ZHAI Honghong, et al. Identification of fiber length related genes using cotton oligonucleotide microarrays[J]. Acta Agronomica Sinica, 2011, 37(1): 95-104.
DOI URL |
|
| [16] | 吴嫚, 李龙云, 裴文锋, 等. 利用马克隆值差异显著的BILs鉴定棉纤维品质相关基因[J]. 棉花学报, 2020, 32(1):52-62. |
| WU Man, LI Longyun, PEI Wenfeng, et al. Identification of differentially expressed genes in developing cotton fibers between two groups of backcross inbred lines differing in fiber Micronaire[J]. Cotton Science, 2020, 32(1):52-62. | |
| [17] |
Hu Y, Chen J D, Fang L, et al. Gossypium barbadense and Gossypium hirsutum genomes provide insights into the origin and evolution of allotetraploid cotton[J]. Nature Genetics, 2019, 51(4):739-748.
DOI URL |
| [18] |
Chee P, Draye X, Jiang C X, et al. Molecular dissection of interspecific variation between Gossypium hirsutum and Gossypium barbadense (cotton) by a backcross-self approach: I. fiber elongation[J]. Theoretical and Applied Genetics, 2005, 111(4):757-763.
DOI URL |
| [19] | 李龙云. 利用基因芯片技术筛选棉纤维发育相关基因[D]. 杨凌: 西北农林科技大学, 2010. |
| LI Longyun. Identification Of Fiber Quality Genes By Using Cotton Oligonucleotide Microarrays with Gene Expression Analysis[D]. Yangling: Northwest A&F University, 2010. | |
| [20] |
Sun Q, Cai Y F, Xie Y F, et al. Gene expression profiling during gland morphogenesis of a mutant and a glandless upland cotton[J]. Molecular Biology Reports, 2010, 37(7):3319-3325.
DOI URL |
| [21] | 佘义斌, 朱一超, 张天真, 等. 棉花腺苷高半胱氨酸水解酶cDNA的克隆、表达及染色体定位[J]. 作物学报, 2008, (6):958-964. |
|
SHE Yibin, ZHU Yichao, ZHANG Tianzhen, et al. Cloning, expression, and mapping of s-adenosyl-l-homocysteine hydrolase (GhSAHH) cDNA in cotton[J]. Acta Agronomica Sinica, 2008, 34(6):958-964.
DOI URL |
|
| [22] |
Li C H, Zhu Y Q, Meng Y L, et al. Isolation of genes preferentially expressed in cotton fibers by cDNA filter arrays and RT-PCR[J]. Plant Science, 2002, 163(6):1113-1120.
DOI URL |
| [23] |
Bach I. The LIM domain: regulation by association[J]. Mechanisms of Development, 2000, 91(1-2):5-17.
PMID |
| [24] |
Saxena I M, Brown R M. Cellulose synthases and related enzymes[J]. Current Opinion in Plant Biology, 2000, 3(6):523-31.
PMID |
| [25] | 武耀廷, 张恒木, 刘进元. 棉纤维细胞发育过程中纤维素的生物合成[J]. 棉花学报, 2003, 15(3):174-179. |
| WU Yaoting, ZHANG Hengmu, LIU Jinyuan. Cellulose biosynthesis in developing cotton fibers[J]. Cotton Science, 2003, 15(3):174-179. | |
| [26] |
Zhang S J, Jiang Z X, Chen J, et al. The cellulose synthase (CesA) gene family in four Gossypium species: phylogenetics, sequence variation and gene expression in relation to fiber quality in Upland cotton[J]. Molecular Genetics and Genomics, 2021, 296(2):355-368.
DOI URL |
| [27] |
Harmer S E, Oxford S J, Timmis J N. Characterization of six alpha-expansin genes in Gossypium hirsutum (upland cotton)[J]. Molecular Genetics and Genomics, 2002, 268(1):1-9.
PMID |
| [28] | Wilkins T A, Arpat A, Sickler B. Cotton fiber genomics: developmental mechanisms[J]. Pflanzenschutz-Nachrichten Bayer, 2005, 1:119-139. |
| [29] |
Gou J Y, Wang L J, Chen S P, et al. Gene expression and metabolite profiles of cotton fiber during cell elongation and secondary cell wall synthesis[J]. Cell Research, 2007, 17(5):422-34.
DOI URL |
| [30] |
Li Y, Tu L L, Pettolino F A, et al. GbEXPATR, a species-specific expansin, enhances cotton fibre elongation through cell wall restructuring[J]. Plant Biotechnology Journal, 2016, 14(3):951-963.
DOI URL |
| [31] | 杨君, 马峙英, 王省芬. 棉花纤维品质改良相关基因研究进展[J]. 中国农业科学, 2016, 49(22):4310-4322,4469-4472. |
| YANG Jun, MA Zhiying, WANG Xingfen. Progress in studies on genes related to fiber quality improvement of cotton[J]. Scientia Agricultura Sinica, 2016, 49(22):4310-4322,4469-4472. | |
| [32] |
Dixon D C, Seagull R W, Triplett B A. Changes in the accumulation of α- and β-tubulin isotypes during cotton fiber development[J]. Plant Physiology, 1994, 105(4):1347-1353.
PMID |
| [33] |
Li X B, Cai L, Cheng N H, et al. Molecular characterization of the cotton GhTUB1 gene that is preferentially expressed in fiber[J]. Plant Physiology, 2002, 130(2), 666-674.
DOI URL |
| [34] | 刘迪秋. 陆地棉纤维特异表达基因的克隆与表达研究[D]. 武汉: 华中农业大学, 2007. |
| LIU Diqiu. Molecular cloning and expression analysis of the genes specifically expressed in fibers of Gossypium hirsutum[D]. Wuhan: Huazhong Agricultural University, 2007 |
| [1] | 张承洁, 胡浩然, 段松江, 吴一帆, 张巨松. 氮肥与密度互作对海岛棉生长发育及产量和品质的影响[J]. 新疆农业科学, 2024, 61(8): 1821-1830. |
| [2] | 李颖, 郭文文, 李江博, 曲延英, 陈全家, 郑凯. 90份转BT基因抗虫棉品种(系)在新疆早熟棉区的适应性评价[J]. 新疆农业科学, 2024, 61(7): 1561-1573. |
| [3] | 巩隽铭, 熊显鹏, 张彩霞, 邵东南, 程帅帅, 孙杰. 陆地棉4-香豆酸辅酶A连接酶基因Gh4CL30的功能分析[J]. 新疆农业科学, 2024, 61(6): 1301-1309. |
| [4] | 马尚洁, 李生梅, 杨涛, 王红刚, 赵康, 庞博, 高文伟. 陆地棉GHWAT1-35基因的克隆及亚细胞定位[J]. 新疆农业科学, 2024, 61(6): 1310-1317. |
| [5] | 何婉洁, 孟涵颖, 支梦婷, 陈静. 双斑长跗萤叶甲雌虫、雄虫触角转录组及差异表达基因分析[J]. 新疆农业科学, 2024, 61(4): 984-995. |
| [6] | 王凯迪, 高晨旭, 裴文锋, 杨书贤, 张文庆, 宋吉坤, 马建江, 王莉, 于霁雯, 陈全家. 陆地棉TRM基因家族的鉴定及纤维品质相关优异单倍型分析[J]. 新疆农业科学, 2024, 61(3): 521-536. |
| [7] | 王伟, 张仁福, 刘海洋, 丁瑞丰, 梁革梅, 姚举. 不同抗性遗传背景棉蚜氟啶虫胺腈及啶虫脒抗性品系转录组分析[J]. 新疆农业科学, 2024, 61(12): 3078-3088. |
| [8] | 崔豫疆, 龚照龙, 王俊铎, 郑巨云, 桑志伟, 阳妮, 梁亚军, 李雪源, 曲延英. 245份陆地棉品种农艺性状及产量构成因素综合评价[J]. 新疆农业科学, 2024, 61(10): 2358-2365. |
| [9] | 黄幸磊, 王为然, 王萌, 朱家辉, 林峰, 秦国礼, 杨静, 阿里甫·艾尔西, 吴全忠, 孔杰. 海岛棉种质资源机采农艺性状相关性分析[J]. 新疆农业科学, 2024, 61(1): 1-8. |
| [10] | 赵康, 任丹, 梁维维, 庞博, 马尚洁, 张梦媛, 高文伟. 陆地棉正反交F2∶3家系主要农艺性状与单株皮棉产量的关系[J]. 新疆农业科学, 2024, 61(1): 19-25. |
| [11] | 王朋, 郑凯, 赵杰银, 高文举, 龙遗磊, 陈全家, 曲延英. 陆地棉种质资源材料的耐热性评价及指标筛选[J]. 新疆农业科学, 2023, 60(9): 2081-2090. |
| [12] | 王辉, 郭金成, 宋佳, 张庭军, 何良荣. 高温胁迫下陆地棉GhCIPK6转基因后代生理生化分析[J]. 新疆农业科学, 2023, 60(9): 2109-2119. |
| [13] | 马青山, 杜霄, 陶志鑫, 韩万里, 龙遗磊, 艾先涛, 胡守林. 陆地棉种质材料机采农艺性状鉴定分析[J]. 新疆农业科学, 2023, 60(8): 1830-1839. |
| [14] | 段松江, 彭增莹, 申莹莹, 木丽迪尔·拜波拉提, 吴一凡, 崔建平, 张巨松. 不同海岛棉品种产量及纤维品质对氮肥的响应[J]. 新疆农业科学, 2023, 60(7): 1569-1579. |
| [15] | 耿翡翡, 孟超敏, 卿桂霞, 周佳敏, 张富厚, 刘逢举. 陆地棉磷高效基因GhMYB4的克隆与表达分析[J]. 新疆农业科学, 2023, 60(6): 1406-1412. |
| 阅读次数 | ||||||
|
全文 |
|
|||||
|
摘要 |
|
|||||