

Xinjiang Agricultural Sciences ›› 2023, Vol. 60 ›› Issue (10): 2590-2600.DOI: 10.6048/j.issn.1001-4330.2023.10.030
• Agricultural Equipment Engineering and Mechanization·Animal Husbandry Veterinarian • Previous Articles
HAN Liyun1,2( ), LI Yanyan2, MA Yun1, HU Honghong1, KANG Xiaolong1, GUO Dianzhi2, SHI Yuangang1(
), LI Yanyan2, MA Yun1, HU Honghong1, KANG Xiaolong1, GUO Dianzhi2, SHI Yuangang1( )
)
Received:2022-12-28
Online:2023-10-20
Published:2023-11-01
Correspondence author:
SHI Yuangang (1962-), male, native place :Yinchuan, Ningxia. professor, research field: animal genetics, breeding and reproduction,(E-mail)Supported by:
韩丽云1,2( ), 李艳艳2, 马云1, 虎红红1, 康小龙1, 郭殿芝2, 史远刚1(
), 李艳艳2, 马云1, 虎红红1, 康小龙1, 郭殿芝2, 史远刚1( )
)
通讯作者:
史远刚(1962-),男,宁夏银川人,教授,博士,博士生导师,研究方向为动物遗传育种与繁殖,(E-mail)作者简介:韩丽云(1989-),女,河北衡水人,硕士研究生,研究方向为动物遗传育种,(E-mail)18152307692@163.com
基金资助:CLC Number:
HAN Liyun, LI Yanyan, MA Yun, HU Honghong, KANG Xiaolong, GUO Dianzhi, SHI Yuangang. A study of association analysis on gene PPARγ, MYF5 and AGPAT5 with body size traits in Xinjiang brown cattle[J]. Xinjiang Agricultural Sciences, 2023, 60(10): 2590-2600.
韩丽云, 李艳艳, 马云, 虎红红, 康小龙, 郭殿芝, 史远刚. 新疆褐牛PPARγ、MYF5和AGPAT5基因SNP突变与体尺性状的关联分析[J]. 新疆农业科学, 2023, 60(10): 2590-2600.
| 基因区域 Gene & SNP | 登陆号 Accession number | 引物序列 Primer sequence (5’-3’) | 扩增片段长度 Product length (bp) |
|---|---|---|---|
| PPARγ-exon7 | NM-181024.2 | F: AATGTGAGTCATTGGCTG | 417 |
| R: GGTGTCAGATTTTTCCCTPHPH | |||
| PPARγ-intron2 | F: CCCGATGGTTGCAGATTA | 1 842 | |
| R: AGGCTCCACTTTGATTGC | |||
| MYF5-exon1 | NM-174116.1 | F:GGCGTCTGCCCTTGTTA | 690 |
| R: TTTGTAAAAGTTGGGAAG | |||
| MYF5-intron1 | F: CTCTGATGGCATGGTAAG | 923 | |
| R: CAGTAGACGATGTCAAAA | |||
| AGPAT5-exon7 | NM_001075932.1 | F: GTAGGGAACTATGGGAGA | 771 |
| R: CACTAACTGCAAAACAACA | |||
| AGPAT5-intron3 | F: TCCCGCTGTATGGCAGTTA | 3 461 | |
| R: CTGGAGTTCCTGCGTTGAT |
Tab.1 Primer sequence
| 基因区域 Gene & SNP | 登陆号 Accession number | 引物序列 Primer sequence (5’-3’) | 扩增片段长度 Product length (bp) |
|---|---|---|---|
| PPARγ-exon7 | NM-181024.2 | F: AATGTGAGTCATTGGCTG | 417 |
| R: GGTGTCAGATTTTTCCCTPHPH | |||
| PPARγ-intron2 | F: CCCGATGGTTGCAGATTA | 1 842 | |
| R: AGGCTCCACTTTGATTGC | |||
| MYF5-exon1 | NM-174116.1 | F:GGCGTCTGCCCTTGTTA | 690 |
| R: TTTGTAAAAGTTGGGAAG | |||
| MYF5-intron1 | F: CTCTGATGGCATGGTAAG | 923 | |
| R: CAGTAGACGATGTCAAAA | |||
| AGPAT5-exon7 | NM_001075932.1 | F: GTAGGGAACTATGGGAGA | 771 |
| R: CACTAACTGCAAAACAACA | |||
| AGPAT5-intron3 | F: TCCCGCTGTATGGCAGTTA | 3 461 | |
| R: CTGGAGTTCCTGCGTTGAT |
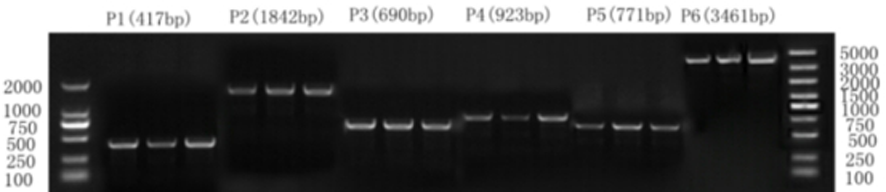
Fig.1 Individual PCR product Note: M represents Marker, P1 represents PPARγ-Exon7, P2 represents PPARγ-Intron2, P3 represents MyF5-Exon1, P4 represents MyF5-Inintron1, P5 represents AGPAT5-Exon7, P6 represents AGPAT5-Inintron3
| 基因 Gene | SNP | 基因型 Genotype | 基因型 频率 Genotypic frequency | 等位基因 Allele | 等位基因 频率 Allele frequency | 纯合度 Homozy- gosity | 杂合度 Heterozy- gosity | 有效 等位 基因数 Effective number of alleles | PIC | 哈代-温伯格平衡 Hardy-weinberg equilibrium | |
|---|---|---|---|---|---|---|---|---|---|---|---|
| x2 | P | ||||||||||
| PPARγ-extron7 | SNP1 | AA(84) | 0.85 | A | 0.92 | 0.86 | 0.14 | 1.16 | 0.13 | 0.66 | 0.42 |
| AB(15) | 0.15 | B | 0.08 | ||||||||
| BB (0) | 0 | ||||||||||
| PPARγ-intron2 | SNP2 | AA(50) | 0.51 | A | 0.70 | 0.58 | 0.42 | 1.74 | 0.33 | 1.62 | 0.20 |
| AB(37) | 0.37 | B | 0.30 | ||||||||
| BB(1) | 0.12 | ||||||||||
| MYF5-exon1 | SNP3 | AA(95) | 0.96 | A | 0.98 | 0.97 | 0.03 | 1.04 | 0.04 | 0.04 | 0.84 |
| AB(4) | 0.04 | B | 0.02 | ||||||||
| BB(0) | 0 | ||||||||||
| MYF5-intron1 | SNP4 | AA(24) | 0.24 | A | 0.62 | 0.63 | 0.47 | 1.89 | 0.36 | 37.56 | <0.001 |
| AB(75) | 0.76 | B | 0.38 | ||||||||
| BB(0) | 0 | ||||||||||
| AGPAT5-intron3 | SNP5 | AA(27) | 0.27 | A | 0.55 | 0.50 | 0.50 | 1.99 | 0.37 | 1.19 | 0.28 |
| AB(54) | 0.55 | B | 0.46 | ||||||||
| BB(18) | 0.18 | ||||||||||
| AGPAT5-exon7 | SNP6 | AA(72) | 0.73 | A | 0.81 | 0.69 | 0.31 | 1.46 | 0.26 | 27.27 | <0.001 |
| AB(15) | 0.15 | B | 0.19 | ||||||||
| BB(12) | 0.12 | ||||||||||
Tab.2 Genotypic and allelic frequencies and Hardy-Weinberg equilibrium test of 6 SNPs
| 基因 Gene | SNP | 基因型 Genotype | 基因型 频率 Genotypic frequency | 等位基因 Allele | 等位基因 频率 Allele frequency | 纯合度 Homozy- gosity | 杂合度 Heterozy- gosity | 有效 等位 基因数 Effective number of alleles | PIC | 哈代-温伯格平衡 Hardy-weinberg equilibrium | |
|---|---|---|---|---|---|---|---|---|---|---|---|
| x2 | P | ||||||||||
| PPARγ-extron7 | SNP1 | AA(84) | 0.85 | A | 0.92 | 0.86 | 0.14 | 1.16 | 0.13 | 0.66 | 0.42 |
| AB(15) | 0.15 | B | 0.08 | ||||||||
| BB (0) | 0 | ||||||||||
| PPARγ-intron2 | SNP2 | AA(50) | 0.51 | A | 0.70 | 0.58 | 0.42 | 1.74 | 0.33 | 1.62 | 0.20 |
| AB(37) | 0.37 | B | 0.30 | ||||||||
| BB(1) | 0.12 | ||||||||||
| MYF5-exon1 | SNP3 | AA(95) | 0.96 | A | 0.98 | 0.97 | 0.03 | 1.04 | 0.04 | 0.04 | 0.84 |
| AB(4) | 0.04 | B | 0.02 | ||||||||
| BB(0) | 0 | ||||||||||
| MYF5-intron1 | SNP4 | AA(24) | 0.24 | A | 0.62 | 0.63 | 0.47 | 1.89 | 0.36 | 37.56 | <0.001 |
| AB(75) | 0.76 | B | 0.38 | ||||||||
| BB(0) | 0 | ||||||||||
| AGPAT5-intron3 | SNP5 | AA(27) | 0.27 | A | 0.55 | 0.50 | 0.50 | 1.99 | 0.37 | 1.19 | 0.28 |
| AB(54) | 0.55 | B | 0.46 | ||||||||
| BB(18) | 0.18 | ||||||||||
| AGPAT5-exon7 | SNP6 | AA(72) | 0.73 | A | 0.81 | 0.69 | 0.31 | 1.46 | 0.26 | 27.27 | <0.001 |
| AB(15) | 0.15 | B | 0.19 | ||||||||
| BB(12) | 0.12 | ||||||||||
| 基因 Gene | 基因型 Genotype | 体长 Length | 胸围 Bust | 胸宽 Chest width | 十字部高 Height at hip cross | 体重 Weight | |
|---|---|---|---|---|---|---|---|
| PPARγ-extron7 | SNP1 | AA | 128.97±1.16 | 158.22±0.89A | 43.30±0.48 | 122.32±0.63 | 300.21±3.86A |
| AB | 129.11±1.28 | 162.07±0.99B | 42.79±0.51 | 122.37±0.71 | 311.76±4.42B | ||
| P值 | 0.921 9 | 0.000 2 | 0.588 5 | 0.957 7 | 0.009 9 | ||
| PPARγ-intron2 | SNP2 | AA | 128.11±1.15 | 160.60±0.88 | 43.43±0.48 | 122.42±0.63 | 313.72±3.87Aa |
| AB | 130.42±1.20 | 161.33±0.94 | 42.79±0.51 | 122.21±0.67 | 396.63±4.11B | ||
| BB | 128.59±1.49 | 158.51±1.15 | 43.21±0.63 | 122.42±0.82 | 307.61±5.02b | ||
| P值 | 0.077 4 | 0.063 1 | 0.324 7 | 0.923 4 | 0.001 9 | ||
| MYF5-exon1 | SNP3 | AA | 128.61±0.89 | 159.51±0.71 | 42.32±0.38 | 121.53±0.50 | 301.65±2.96 |
| AB | 129.47±1.88 | 160.79±1.42 | 43.97±0.78 | 123.17±1.01 | 310.32±6.28 | ||
| P值 | 0.681 8 | 0.421 8 | 0.061 2 | 0.147 9 | 0.208 7 | ||
| MYF5-intron1 | SNP4 | AA | 128.93±1.24 | 159.89±0.97 | 42.59±0.53a | 122.75±0.69 | 301.65±2.96 |
| AB | 129.15±1.06 | 160.40±0.80 | 43.70±0.44b | 121.95±0.57 | 310.32±6.28 | ||
| P值 | 0.832 2 | 0.549 8 | 0.014 2 | 0.176 2 | 0.588 7 | ||
| AGPAT5-intron3 | SNP5 | AA | 129.19±1.26 | 160.96±0.96A | 43.58±0.53A | 123.17±0.68 | 313.60±4.25A |
| AB | 130.54±1.17a | 161.99±0.90A | 43.69±0.49A | 122.36±0.64 | 310.47±3.96A | ||
| BB | 127.39±1.33b | 157.50±1.04B | 42.16±0.56B | 121.52±0.74 | 293.88±4.49B | ||
| P值 | 0.039 5 | 0.001 7 | 0.008 6 | 0.088 0 | 0.000 3 | ||
| AGPAT5-exon7 | SNP6 | AA | 128.81±1.18 | 161.35±0.91A | 42.92±0.50a | 123.06±0.65 | 301.86±3.97 |
| AB | 130.02±1.29 | 162.91±0.98A | 44.14±0.54b | 122.86±0.69 | 311.17±4.38 | ||
| BB | 128.30±1.55 | 156.18±1.22B | 42.36±0.62a | 121.12±0.86 | 304.89±5.21 | ||
| P值 | 0.523 9 | 0.000 2 | 0.019 5 | 0.076 1 | 0.093 7 |
Tab.3 Effects of 6 SNPs on body size in Xinjiang cattle(LSE±SE)
| 基因 Gene | 基因型 Genotype | 体长 Length | 胸围 Bust | 胸宽 Chest width | 十字部高 Height at hip cross | 体重 Weight | |
|---|---|---|---|---|---|---|---|
| PPARγ-extron7 | SNP1 | AA | 128.97±1.16 | 158.22±0.89A | 43.30±0.48 | 122.32±0.63 | 300.21±3.86A |
| AB | 129.11±1.28 | 162.07±0.99B | 42.79±0.51 | 122.37±0.71 | 311.76±4.42B | ||
| P值 | 0.921 9 | 0.000 2 | 0.588 5 | 0.957 7 | 0.009 9 | ||
| PPARγ-intron2 | SNP2 | AA | 128.11±1.15 | 160.60±0.88 | 43.43±0.48 | 122.42±0.63 | 313.72±3.87Aa |
| AB | 130.42±1.20 | 161.33±0.94 | 42.79±0.51 | 122.21±0.67 | 396.63±4.11B | ||
| BB | 128.59±1.49 | 158.51±1.15 | 43.21±0.63 | 122.42±0.82 | 307.61±5.02b | ||
| P值 | 0.077 4 | 0.063 1 | 0.324 7 | 0.923 4 | 0.001 9 | ||
| MYF5-exon1 | SNP3 | AA | 128.61±0.89 | 159.51±0.71 | 42.32±0.38 | 121.53±0.50 | 301.65±2.96 |
| AB | 129.47±1.88 | 160.79±1.42 | 43.97±0.78 | 123.17±1.01 | 310.32±6.28 | ||
| P值 | 0.681 8 | 0.421 8 | 0.061 2 | 0.147 9 | 0.208 7 | ||
| MYF5-intron1 | SNP4 | AA | 128.93±1.24 | 159.89±0.97 | 42.59±0.53a | 122.75±0.69 | 301.65±2.96 |
| AB | 129.15±1.06 | 160.40±0.80 | 43.70±0.44b | 121.95±0.57 | 310.32±6.28 | ||
| P值 | 0.832 2 | 0.549 8 | 0.014 2 | 0.176 2 | 0.588 7 | ||
| AGPAT5-intron3 | SNP5 | AA | 129.19±1.26 | 160.96±0.96A | 43.58±0.53A | 123.17±0.68 | 313.60±4.25A |
| AB | 130.54±1.17a | 161.99±0.90A | 43.69±0.49A | 122.36±0.64 | 310.47±3.96A | ||
| BB | 127.39±1.33b | 157.50±1.04B | 42.16±0.56B | 121.52±0.74 | 293.88±4.49B | ||
| P值 | 0.039 5 | 0.001 7 | 0.008 6 | 0.088 0 | 0.000 3 | ||
| AGPAT5-exon7 | SNP6 | AA | 128.81±1.18 | 161.35±0.91A | 42.92±0.50a | 123.06±0.65 | 301.86±3.97 |
| AB | 130.02±1.29 | 162.91±0.98A | 44.14±0.54b | 122.86±0.69 | 311.17±4.38 | ||
| BB | 128.30±1.55 | 156.18±1.22B | 42.36±0.62a | 121.12±0.86 | 304.89±5.21 | ||
| P值 | 0.523 9 | 0.000 2 | 0.019 5 | 0.076 1 | 0.093 7 |
| 位点 Site | 单倍型 Haplt- type | 单倍型频率 Haplotype frequency | 体长 Length | 胸围 Bust | 胸宽 Chest width | 十字部高 Height at hip cross | 体重 Weight |
|---|---|---|---|---|---|---|---|
| PPARγ- exon7(74) -intron2 (133、170) | GGC | 0.417 7 | 129.51±0.82 | 161.81±0.81 | 43.51±0.48a | 122.23±0.61 | 304.67±3.02 |
| GCC | 0.316 5 | 127.62±1.38 | 161.83±1.36 | 44.50±0.80A | 123.91±1.01a | 306.55±4.99 | |
| GGT | 0.113 9 | 129.93±0.72 | 161.17±0.68 | 43.97±0.41A | 122.63±0.51 | 307.77±2.61 | |
| GCT | 0.113 9 | 129.87±1.40 | 159.68±1.33 | 41.14±0.80Bb | 124.47±1.00a | 308.62±4.99 | |
| TCT | 0.038 0 | 131.60±2.35 | 162.89±2.27 | 45.50±1.37A | 119.18±1.70b | 298.14±8.64 |
Tab.4 Effects of haplotype block PPARγ-exon7(74)-intron2(133、170) on body size traits in Xinjiang brown cattle (LSE±SE)
| 位点 Site | 单倍型 Haplt- type | 单倍型频率 Haplotype frequency | 体长 Length | 胸围 Bust | 胸宽 Chest width | 十字部高 Height at hip cross | 体重 Weight |
|---|---|---|---|---|---|---|---|
| PPARγ- exon7(74) -intron2 (133、170) | GGC | 0.417 7 | 129.51±0.82 | 161.81±0.81 | 43.51±0.48a | 122.23±0.61 | 304.67±3.02 |
| GCC | 0.316 5 | 127.62±1.38 | 161.83±1.36 | 44.50±0.80A | 123.91±1.01a | 306.55±4.99 | |
| GGT | 0.113 9 | 129.93±0.72 | 161.17±0.68 | 43.97±0.41A | 122.63±0.51 | 307.77±2.61 | |
| GCT | 0.113 9 | 129.87±1.40 | 159.68±1.33 | 41.14±0.80Bb | 124.47±1.00a | 308.62±4.99 | |
| TCT | 0.038 0 | 131.60±2.35 | 162.89±2.27 | 45.50±1.37A | 119.18±1.70b | 298.14±8.64 |
| 位点 Site | 单倍型 Haplt- type | 单倍型频率 Haplotype frequency | 体长 Length | 胸围 Bust | 胸宽 Chest width | 十字部高 Height at hip cross | 体重 Weight |
|---|---|---|---|---|---|---|---|
| MYF5- exon1(236) -intron1 (633) | CG | 0.021 9 | 129.43±2.91a | 170.47±2.87A | 46.45±1.65 | 121.64±2.03 | 349.81±1.24A |
| TA | 0.252 7 | 128.31±0.88 | 161.60±0.83B | 44.50±0.48a | 120.64±0.59A | 300.99±3.31B | |
| TG | 0.725 2 | 123.50±0.53b | 161.25±0.51B | 43.31±0.30b | 123.13±0.36B | 307.18±1.96B |
Tab.5 Effects of haplotype block MYF5-exon1(236)-intron1(633) on body size traits in Xinjiang brown cattle(LSE±SE)
| 位点 Site | 单倍型 Haplt- type | 单倍型频率 Haplotype frequency | 体长 Length | 胸围 Bust | 胸宽 Chest width | 十字部高 Height at hip cross | 体重 Weight |
|---|---|---|---|---|---|---|---|
| MYF5- exon1(236) -intron1 (633) | CG | 0.021 9 | 129.43±2.91a | 170.47±2.87A | 46.45±1.65 | 121.64±2.03 | 349.81±1.24A |
| TA | 0.252 7 | 128.31±0.88 | 161.60±0.83B | 44.50±0.48a | 120.64±0.59A | 300.99±3.31B | |
| TG | 0.725 2 | 123.50±0.53b | 161.25±0.51B | 43.31±0.30b | 123.13±0.36B | 307.18±1.96B |
| 位点 Site | 单倍型 Haplt- type | 单倍型频率 Haplotype frequency | 体长 Length | 胸围 Bust | 胸宽 Chest width | 十字部高 Height at hip cross | 体重 Weight |
|---|---|---|---|---|---|---|---|
| AGPAT5- exon7 (72、97)- intron3 (501、515、 550) | AAGGG | 0.062 5 | 133.25±2.14Aa | 160.43±2.15A | 45.41±1.10A | 116.57±1.30B | 316.37±8.14A |
| AGCTT | 0.062 5 | 123.39±1.98B | 149.60±2.06B | 37.52±1.04B | 120.60±1.25A | 274.02±8.14C | |
| ATGGG | 0.062 5 | 133.25±2.14Ab | 160.43±2.15A | 45.40±1.10A | 116.57±1.30B | 316.37±8.14A | |
| GGGGG | 0.312 5 | 130.99±0.82Ab | 160.83±0.84Aa | 44.08±0.43Aa | 122.20±0.51A | 296.26±3.28B | |
| GTCTT | 0.390 6 | 129.22±1.64b | 161.31±1.70A | 43.07±0.82Aa | 123.06±1.00A | 305.69±6.15B | |
| GTGGG | 0.109 4 | 127.75±0.94Ab | 164.20±0.96Ab | 45.49±0.49b | 121.89±0.59A | 321.68±3.64A |
Tab.6 Effects of haplotype block AGPAT5-exon7(72、97)-intron3(501、515、550) on body size traits in brown cattle(LSE±SE)
| 位点 Site | 单倍型 Haplt- type | 单倍型频率 Haplotype frequency | 体长 Length | 胸围 Bust | 胸宽 Chest width | 十字部高 Height at hip cross | 体重 Weight |
|---|---|---|---|---|---|---|---|
| AGPAT5- exon7 (72、97)- intron3 (501、515、 550) | AAGGG | 0.062 5 | 133.25±2.14Aa | 160.43±2.15A | 45.41±1.10A | 116.57±1.30B | 316.37±8.14A |
| AGCTT | 0.062 5 | 123.39±1.98B | 149.60±2.06B | 37.52±1.04B | 120.60±1.25A | 274.02±8.14C | |
| ATGGG | 0.062 5 | 133.25±2.14Ab | 160.43±2.15A | 45.40±1.10A | 116.57±1.30B | 316.37±8.14A | |
| GGGGG | 0.312 5 | 130.99±0.82Ab | 160.83±0.84Aa | 44.08±0.43Aa | 122.20±0.51A | 296.26±3.28B | |
| GTCTT | 0.390 6 | 129.22±1.64b | 161.31±1.70A | 43.07±0.82Aa | 123.06±1.00A | 305.69±6.15B | |
| GTGGG | 0.109 4 | 127.75±0.94Ab | 164.20±0.96Ab | 45.49±0.49b | 121.89±0.59A | 321.68±3.64A |
| [1] |
张青青, 杨洋, 张鸣, 等. 贵州地方黄牛不同组织中PPARγ基因表达水平研究[J]. 中国畜牧兽医, 2017, 44(1): 38-43.
DOI |
| ZHANG Qingqing, YANG Yang, ZHANG Ming. The Expression Level of PPARγ Gene in Different Tissues of Guizhou Cattle Resources[J]. China Animal Husbandry & Veterinary Medicine, 2017, 44(1): 38-43. | |
| [2] |
Abdelrahman M, Sivarajah A, Thiemermann C. Beneficial effects of PPAR-γ ligands in ischemia-reperfusion injury, inflammation and shock[J]. Cardiovascular Research, 2005, 65(4): 772-781.
DOI PMID |
| [3] | Montanari R, Capelli D, Yamamoto K, et al. Insights into PPARγ Phosphorylation and Its Inhibition Mechanism[J]. Journal of Medicinal Chemistry, 2020. |
| [4] |
Pownall M E, Gustafsson M K, Emerson C P. Myogenic regulatory factors and the specification of muscle progenitors in vertebrate embryos[J]. Annu Rev Cell Dev Biol, 2002, 18(1): 747-783.
DOI URL |
| [5] |
Weintraub H, Davis R, Tapscott S, et al. The myoD gene family: nodal point during specification of the muscle cell lineage[J]. Science, 1991, 251(4995): 761-766.
PMID |
| [6] | Zammit P S, Carvajal J J, Golding J P, et al. Myf5 expression in satellite cells and spindles in adult muscle is controlled by separate genetic elements[J]. Developmental Biology, 2004, 273(2). |
| [7] |
Cz A, Shar A, Rk A, et al. Genetic variants in MYF5 affected growth traits and beef quality traits in Chinese Qinchuan cattle-Science Direct[J]. Genomics, 2020, 112(4): 2804-2812.
DOI PMID |
| [8] |
Wang J, Hu Y, Elzo M A, et al. Genetic effect of Myf5 gene in rabbit meat quality traits[J]. Journal of Genetics, 2017, 96(4): 673-679.
PMID |
| [9] | Ye G M, Chen C, Huang S, et al. Cloning and characterization a novel human 1-acyl-sn-glycerol-3-phosphate acyltransferase gene AGPAT7[J]. Dna Seq, 2005, 16(5): 386-390. |
| [10] | Takeuchi K, Reue K. Biochemistry, physiology, and genetics of GPAT, AGPAT, and lipin enzymes in triglyceride synthesis[J]. American Journal of Physiology - Endocrinology and Metabolism, 2009, 296(6). |
| [11] | Prasad S S, Garg A, Agarwal A K. Enzymatic activities of the human AGPAT isoform 3 and isoform 5: localization of AGPAT5 to mitochondria[J]. Journal of Lipid Research, 2011, 52(3). |
| [12] | 卫喜明, 张金玉, 孙晓玉, 等. 德系西门塔尔牛AGPAT5基因多态性及其与肉质性状的相关分析[J]. 黑龙江畜牧兽医, 2013,(9): 38-40. |
| WEI Ximing, ZHANG Jinyu, SUN Xiaoyu. Correlation analysis between the polymorphism of AGPAT5 gene and meat quality traits in German Simmental cattle[J]. HeilongjiangAnimal Husbandry & Veterinary Medicine, 2013,(9): 38-40. | |
| [13] | Edea Z, Jung K S, Shin S S, et al. Signatures of positive selection underlying beef production traits in Korean cattle breeds[J]. Journal of Animal Science and Technology, 2020. |
| [14] | 曲永利, 陈勇. 养牛学[M]. 北京: 北京工业出版社, 2014. |
| QU Yongli, CHEN Yong. Cattle Science[M]. Beijing: Beijing Industrial Press, 2014. | |
| [15] |
Orozco G, Hinks A, Eyre S, et al. Combined effects of three independent SNPs greatly increase the risk estimate for RA at 6q23[J]. Human Molecular Genetics, 2009, 18(14): 2693.
DOI PMID |
| [16] | Chen X, Raza S, Cheng G, et al. Bta-miR-376a Targeting KLF15 Interferes with Adipogenesis Signaling Pathway to Promote Differentiation of Qinchuan Beef Cattle Preadipocytes[J]. Animals, 2020, 10(12). |
| [17] | Al-Ghadban S, Diaz Z T, Singer H J, et al. Increase in Leptin and PPAR-γ Gene Expression in Lipedema Adipocytes Differentiated in vitro from Adipose-Derived Stem Cells[J]. Cells 2020, 9(2). |
| [18] | 李丽, 徐琪, 李柳萌, 等. PPARγ基因多态性与绍兴鸭生长发育性能关联分析[J]. 扬州大学学报(农业与生命科学版), 2015, 36(4): 35-39. |
| LI Li, XU Qi, LI Liumeng. The correlation between polymorphism ofPPARγ genes with the performance of growth and slaughter in Shaoxing ducks[J]. Journal of Yangzhou University (Agriculture and Life Sciences Ed.), 2015, 36(4): 35-39. | |
| [19] | 谢相林, 王栋, 王慧, 等. PPARα基因多态性与肉鸡脂肪性状的相关性分析[J]. 畜牧兽医学报, 2005, 36(12): 1261-1264. |
| XIE Xianglin, WANG Dong, WANG Hui, et al. Relationship between Genotype of PPARα and Body Fat Traits in AA Broiler Line[J]. Acta Veterinaria Et Zootechnica Sinica, 2005, 36(12),1261-1264. | |
| [20] | 纪永吉. 牦牛PPARα、PPARγ、PPARδ基因的多态性与生长性状相关性研究[D]. 兰州: 甘肃农业大学. 2016 |
| JI Yongji. Study on correlation between single nucleotide polymorphism of PPARα,PPARγ,PPARδ gene and growth traits in Yak[D]. Lanzhou: Gansu Agricultural University, 2016 | |
| [21] | 孙太红. 牛PPARα、PPARγ基因多态性及其与生长性状相关性分析[D]. 信阳: 信阳师范学院, 2015. |
| SUN Taihong. The analysis of PPARα, PPARγ gene polymorphisms in bovine and their association with growth traits[D]. Xinyang: Xinyang Normal University, 2015. | |
| [22] |
Min L, Jian P, Xu D Q, et al. Association of MYF5 and MYOD1 gene polymorphisms and meat quality traits in Large White x Meishan F2 pig populations[J]. Biochemical Genetics, 2008, 46(s11-12): 720-732.
DOI URL |
| [23] |
Cz A, Shar A, Rk A, et al. Genetic variants in MYF5 affected growth traits and beef quality traits in Chinese Qinchuan cattle - Science Direct[J]. Genomics, 2020, 112(4): 2804-2812.
DOI PMID |
| [24] |
Yin H, Zhang Z, Xi L, et al. Association of MyF5, MyF6 and MyOG Gene Polymorphisms with Carcass Traits in Chinese Meat Type Quality Chicken Populations[J]. Journal of Animal and Veterinary Advances,. 2011, 10(6): 704-708.
DOI URL |
| [25] | 姚毅, 马红艳, 吕学斌, 等. Myf5基因多态性与山羊生长性能的相关性分析[J]. 山西农业科学, 2015, 43(12):1672-1675. |
| YAO Yi, MA Hongyan, LYU Xuebin. Association of Polymorphisms of Myf 5 Gene with Growth Performances in Goats[J]. Journal of Shanxi Agricultural Sciences, 2015, 43(12):1672-1675. | |
| [26] | 卫喜明. 不同品种牛肉用性能及肉质性状相关基因分析[D]. 长春: 吉林大学, 2013. |
| Wei Ximing. Meat Performance and Correction analysis with Meat Quality Traits in different varieties of cattle[D]. Changchun: Jilin University, 2013. |
| [1] | SU Nan, MA Zen, WANG Xiao, CHEN Wenzhong, ZHANG Yuxia, LIU Wujun, YAN Xiangming. Study on the differences of slaughter performance and meat quality characteristics of Xinjiang Brown Cattle by different feeding methods [J]. Xinjiang Agricultural Sciences, 2024, 61(12): 3105-3112. |
| [2] | TANG Li, TIAN Kechuan, ZHANG Xinning, LIU Li, Abulikemu Adili, YANG Zhi, YANG Cunming, ZHANG Xiaoxue, HUANG Xixia, TIAN Yuezhen. Clustering and principal component analysis of Hotan sheep body weight indexes in different growth stages [J]. Xinjiang Agricultural Sciences, 2024, 61(11): 2853-2860. |
| [3] | WEI Yong, GAO Yu, YANG Min, XU Zihao, REN Wanping. Effects of feeding steam-pressed corn on meat performance and liver nutrient metabolism of Xinjiang brown cattle [J]. Xinjiang Agricultural Sciences, 2023, 60(10): 2583-2589. |
| [4] | DU Wei, LI Hongbo, ZHOU Zhenyong, ZHANG Jinshan, CUI Fanrong, YE Zhibing, YUAN Lixing, ZHANG YANG. Comparative Analysis on Meat Performance and Meat Quality of Xinjiang Brown Cattle and Kazak Cattle [J]. Xinjiang Agricultural Sciences, 2022, 59(7): 1814-1819. |
| [5] | LI Chao, LI Xiaobin, DENG Haifeng, HUANG Xinxin, YANG Fan, LI Hai, CHEN Kaixu. Study on the Changes of Body Weight and Body Size Increase of 0-12 Months Old Thoroughbred Horse Foal [J]. Xinjiang Agricultural Sciences, 2022, 59(6): 1545-1552. |
| [6] | LI Hongbo, LI Na, YAN Xiangmin, YE Zhibing, CUI Fanrong, ZHANG Yang. Correlation between Leptin Gene Expression Level in Different Tissues and Meat Quality in Xinjiang Brown Cattle [J]. Xinjiang Agricultural Sciences, 2022, 59(4): 1034-1040. |
| [7] | DU Wei, ZHANG Jinshan, LI Hongbo, ZHOU Zhenyong, CUI Fanrong, YE Zhibing, ZHANG Yang. Production Performance Measurement and Breeding Improvement for Xinjiang Brown Cattle(Meat Type) [J]. Xinjiang Agricultural Sciences, 2021, 58(5): 973-980. |
| [8] | WANG Jian-wen;ZENG Ya-qi;YAO Xin-kui;MENG Jun;LIU Jia;Cairendaoerji;ZHANG Kai-li. Fitting the Body Size Growth Curve of Yanqi Stallion [J]. , 2016, 53(8): 1546-1553. |
| [9] | ZHOU Zhen-yong;LI Na;LI Hong-bo;YAN Xiang-min;ZHANG Jin-shan;DU Wei;ZHANG Yang. Population Genetic Diversity of Xinjiang Brown Cattle [J]. , 2016, 53(7): 1356-1363. |
| [10] | . Studies on the Comprehensive Evaluation Model of Young Yili Horse Body Size [J]. , 2016, 53(4): 771-777. |
| [11] | . Study on Growth Curve Fitting of Xinjiang Brown Cattle and Their Hybrids [J]. , 2016, 53(3): 569-574. |
| [12] | . Analysis of Influencing Factors on Xinjiang Brown Cattle and Holstein Cattle in Pregnancy Time [J]. , 2016, 53(10): 1947-1953. |
| [13] | . Change, Correlation and Regression Analysis of Body Weight and Body Size Index in Kyrgyz Mare at Different Ages [J]. , 2014, 51(10): 1942-1947. |
| [14] | GU Wei-rong;SUN Zong-jiu;ZHU Jin-zhong. Influence of Different Grazing Gradients on Xinjiang Brown Cattle Grazing Behavior [J]. , 2013, 50(5): 944-948. |
| [15] | LI Na;LI Hong-bo;YAN Xiang-min;ZHANG Jin-shan;ZHOU Zhen-yong;YAO Gang;ZHANG Yang. Comparative Analysis of the Slaughter Performance and Beef Quality between Xinjiang Brown Cattle and Other Dual Purpose Cattle [J]. , 2013, 50(12): 2317-2324. |
| Viewed | ||||||||||||||||||||||||||||||||||||||||||||||||||
|
Full text 57
|
|
|||||||||||||||||||||||||||||||||||||||||||||||||
|
Abstract 209
|
|
|||||||||||||||||||||||||||||||||||||||||||||||||