

新疆农业科学 ›› 2025, Vol. 62 ›› Issue (5): 1084-1091.DOI: 10.6048/j.issn.1001-4330.2025.05.005
丁树根1( ), 石玉杰1, 阿布都克尤木·阿不都热孜克2, 徐麟2, 吴元龙1, 李志博1, 林海荣1, 赵曾强3, 聂新辉1(
), 石玉杰1, 阿布都克尤木·阿不都热孜克2, 徐麟2, 吴元龙1, 李志博1, 林海荣1, 赵曾强3, 聂新辉1( )
)
收稿日期:2024-10-11
出版日期:2025-05-20
发布日期:2025-07-09
通信作者:
聂新辉(1982-),女,新疆人,教授,博士,硕士生/博士生导师,研究方向为棉花分子育种,(E-mail)xjnxh2004130@126.com作者简介:丁树根(1998-),男,安徽人,硕士研究生,研究方向为棉花作物遗传育种,(E-mail)dsg199878@gmail.com
基金资助:
DING Shugen1( ), SHI Yujie1, Abudukeyoumu Abudurezike2, XU Lin2, WU Yuanlong1, LI Zhibo1, LIN Hairong1, ZHAO Zengqiang3, NIE Xinhui1(
), SHI Yujie1, Abudukeyoumu Abudurezike2, XU Lin2, WU Yuanlong1, LI Zhibo1, LIN Hairong1, ZHAO Zengqiang3, NIE Xinhui1( )
)
Received:2024-10-11
Published:2025-05-20
Online:2025-07-09
Supported by:摘要:
【目的】基于SSR(Simple sequence repeat,简单重复序列)标记对棉花SOC(Seed oil content,种子油分含量)进行关联分析,挖掘优异等位变异位点,解析棉花种子油分含量性状的遗传机理,为棉花高油分育种提供理论参考。【方法】利用筛选出覆盖棉花全基因组的145对SSR标记对245份棉花材料进行多态性扫描;利用R语言绘制群体表型分布和相关关系图,采用TASSEL软件的混合线性模型进行关联分析,挖掘与SOC相关的优异等位变异位点。【结果】获得34个与SOC相关的等位变异位点(P<0.05)。表型变异解释率为1.83%~8.7%,平均值为5.70%。【结论】挖掘到34个与油分含量性状相关等位变异位点,以及棉花纤维品质和产量相关的位点9个。
中图分类号:
丁树根, 石玉杰, 阿布都克尤木·阿不都热孜克, 徐麟, 吴元龙, 李志博, 林海荣, 赵曾强, 聂新辉. 棉花种子油分含量QTL的定位及遗传效应分析[J]. 新疆农业科学, 2025, 62(5): 1084-1091.
DING Shugen, SHI Yujie, Abudukeyoumu Abudurezike, XU Lin, WU Yuanlong, LI Zhibo, LIN Hairong, ZHAO Zengqiang, NIE Xinhui. Mapping and genetic effect analysis of QTL for cotton seed oil content[J]. Xinjiang Agricultural Sciences, 2025, 62(5): 1084-1091.
| 环境 Environments | 最小值 Min(%) | 最大值 Max(%) | 均值 Mean(%) | 标准差 SD | 变异系数 CV(%) | 广义遗传力 H2(%) |
|---|---|---|---|---|---|---|
| 2018年库尔勒 Korla in 2018 | 13.41 | 21.98 | 17.70 | 3.29 | 5.38 | 96.98 |
| 2019年库尔勒 Korla in 2019 | 14.45 | 21.76 | 17.79 | 1.93 | 9.22 | |
| 2018年石河子 Shihezi 2018 | 13.96 | 21.05 | 17.43 | 1.86 | 9.37 | |
| 2019年石河子 Shihezi 2019 | 14.17 | 19.67 | 16.84 | 1.59 | 10.59 |
表1 油分性状在4个环境下的表型变异
Tab.1 Phenotypic variation of oil traits in four environments
| 环境 Environments | 最小值 Min(%) | 最大值 Max(%) | 均值 Mean(%) | 标准差 SD | 变异系数 CV(%) | 广义遗传力 H2(%) |
|---|---|---|---|---|---|---|
| 2018年库尔勒 Korla in 2018 | 13.41 | 21.98 | 17.70 | 3.29 | 5.38 | 96.98 |
| 2019年库尔勒 Korla in 2019 | 14.45 | 21.76 | 17.79 | 1.93 | 9.22 | |
| 2018年石河子 Shihezi 2018 | 13.96 | 21.05 | 17.43 | 1.86 | 9.37 | |
| 2019年石河子 Shihezi 2019 | 14.17 | 19.67 | 16.84 | 1.59 | 10.59 |
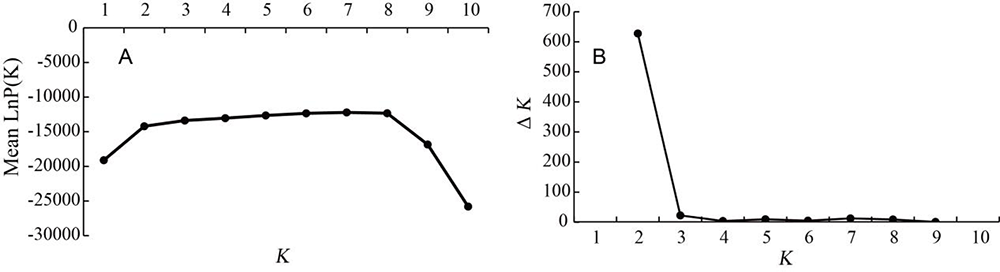
图2 基于 Structure 分析 K 值与 ln(P(D))值和 ΔK 值折线 注:A:K 值与 Mean lnP(K)值折线图;B:ΔK 值随 K 值变化
Fig.2 Lines graph of K value with ln(P(D)) value and ΔK value based on structure analysis Notes:A: Line chart of K value and Mean lnP(K); B: Line chart of ΔK changing with K values
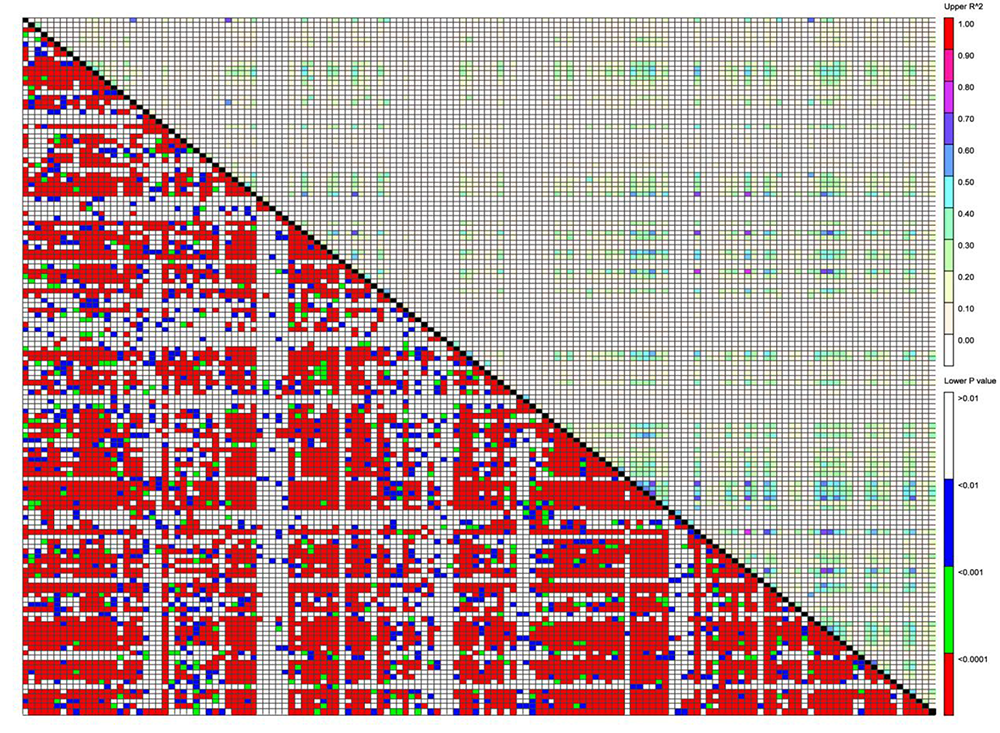
图4 自然群体棉花品种 26 条染色体145个SSR标记位点间的连锁不平衡分布
Fig.4 Linkage disequilibrium distribution among 145 SSR marker loci on 26 chromosomes of natural population cotton varieties
| 性状 Trait | 位点 Locus | P_FDR | 表型变 异解释率 Phenotypic variation explanatiory rateR2(%) |
|---|---|---|---|
| HAU4483b** | 2.38E-06 | 8.70 | |
| HAU4483a** | 6.10E-06 | 8.68 | |
| MON_SHIN-1585a** | 6.29E-06 | 7.54 | |
| MON_SHIN-1481a** | 1.14E-05 | 7.32 | |
| MON_CGR5447a** | 1.96E-05 | 7.00 | |
| HAU4022b** | 8.97E-05 | 6.93 | |
| MUSS422aa** | 1.45E-04 | 6.90 | |
| SOC | HAU3071ad** | 3.18E-04 | 6.82 |
| 种子油分含量 | HAU0875** | 0.0034 | 6.69 |
| NAU3346ba** | 0.0036 | 6.65 | |
| MON_DPL0375ca** | 0.0042 | 6.61 | |
| HAU-SNP113b** | 0.0055 | 6.54 | |
| NAU3774a** | 0.0061 | 6.54 | |
| NAU3736bb* | 0.0241 | 6.38 | |
| HAU2588a* | 0.0285 | 6.37 | |
| HAU2588b* | 0.0368 | 6.36 | |
| NAU3827b* | 0.0395 | 6.30 | |
| MUSS422ab* | 0.0432 | 6.06 | |
| NAU5172b* | 0.029 | 5.18 | |
| HAU-SNP113a* | 0.0362 | 5.14 | |
| NBRI_HQ526730c* | 0.0422 | 5.00 | |
| NBRI_HQ527566* | 0.0121 | 5.00 | |
| DPL0062* | 0.0183 | 4.98 | |
| HAU4022c* | 0.0224 | 4.84 | |
| SOC | BNL2449b* | 0.0237 | 4.83 |
| 种子油分含量 | BNL2449a* | 0.029 | 4.76 |
| HAU1952bc* | 0.0368 | 4.71 | |
| NAU7182* | 0.0395 | 4.55 | |
| NAU3774b* | 0.0432 | 4.47 | |
| MON_DPL0893a* | 0.0493 | 4.46 | |
| BNL2535ba* | 0.0496 | 4.45 | |
| BNL2535bb** | 1.14E-05 | 3.06 | |
| NBRI_HQ527820a** | 1.96E-05 | 2.06 | |
| MON_DPL0502a** | 8.97E-05 | 1.83 |
表2 与种子油分含量关联的SSR位点
Tab.2 SSR locus associated with seed oil content
| 性状 Trait | 位点 Locus | P_FDR | 表型变 异解释率 Phenotypic variation explanatiory rateR2(%) |
|---|---|---|---|
| HAU4483b** | 2.38E-06 | 8.70 | |
| HAU4483a** | 6.10E-06 | 8.68 | |
| MON_SHIN-1585a** | 6.29E-06 | 7.54 | |
| MON_SHIN-1481a** | 1.14E-05 | 7.32 | |
| MON_CGR5447a** | 1.96E-05 | 7.00 | |
| HAU4022b** | 8.97E-05 | 6.93 | |
| MUSS422aa** | 1.45E-04 | 6.90 | |
| SOC | HAU3071ad** | 3.18E-04 | 6.82 |
| 种子油分含量 | HAU0875** | 0.0034 | 6.69 |
| NAU3346ba** | 0.0036 | 6.65 | |
| MON_DPL0375ca** | 0.0042 | 6.61 | |
| HAU-SNP113b** | 0.0055 | 6.54 | |
| NAU3774a** | 0.0061 | 6.54 | |
| NAU3736bb* | 0.0241 | 6.38 | |
| HAU2588a* | 0.0285 | 6.37 | |
| HAU2588b* | 0.0368 | 6.36 | |
| NAU3827b* | 0.0395 | 6.30 | |
| MUSS422ab* | 0.0432 | 6.06 | |
| NAU5172b* | 0.029 | 5.18 | |
| HAU-SNP113a* | 0.0362 | 5.14 | |
| NBRI_HQ526730c* | 0.0422 | 5.00 | |
| NBRI_HQ527566* | 0.0121 | 5.00 | |
| DPL0062* | 0.0183 | 4.98 | |
| HAU4022c* | 0.0224 | 4.84 | |
| SOC | BNL2449b* | 0.0237 | 4.83 |
| 种子油分含量 | BNL2449a* | 0.029 | 4.76 |
| HAU1952bc* | 0.0368 | 4.71 | |
| NAU7182* | 0.0395 | 4.55 | |
| NAU3774b* | 0.0432 | 4.47 | |
| MON_DPL0893a* | 0.0493 | 4.46 | |
| BNL2535ba* | 0.0496 | 4.45 | |
| BNL2535bb** | 1.14E-05 | 3.06 | |
| NBRI_HQ527820a** | 1.96E-05 | 2.06 | |
| MON_DPL0502a** | 8.97E-05 | 1.83 |
| 位点 Locus | 染色体 Chr | 物理位置 Location (bp) | 已报道研究 性状及文献 Reported traits and references |
|---|---|---|---|
| HAU3071 | Chr14 | 307 | FE[ |
| NAU3346 | Chr15 | 305 | FL[ |
| NAU3736 | Chr01 | 203 | FE[ |
| NAU3827 | Chr18 | 136 | LY[ |
| NAU3774 | Chr26 | 293 | FU[ MV[ |
| DPL0062 | Chr21 | 137 | LW[ FU[ |
| MUSS422 | Chr01 | 207 | FU[ SFC[ |
| BNL2449 | Chr13 | 183 | BW[ MV[ LP[ |
| NBRI_HQ526730 | Chr01 | 228 | SI[ |
表3 试验研究与已报道位点的比较结果
Tab.3 Comparative results of this study with reported sites
| 位点 Locus | 染色体 Chr | 物理位置 Location (bp) | 已报道研究 性状及文献 Reported traits and references |
|---|---|---|---|
| HAU3071 | Chr14 | 307 | FE[ |
| NAU3346 | Chr15 | 305 | FL[ |
| NAU3736 | Chr01 | 203 | FE[ |
| NAU3827 | Chr18 | 136 | LY[ |
| NAU3774 | Chr26 | 293 | FU[ MV[ |
| DPL0062 | Chr21 | 137 | LW[ FU[ |
| MUSS422 | Chr01 | 207 | FU[ SFC[ |
| BNL2449 | Chr13 | 183 | BW[ MV[ LP[ |
| NBRI_HQ526730 | Chr01 | 228 | SI[ |
| [1] | 许红霞, 杨伟华, 王延琴, 等. 我国油用棉子质量状况分析[J]. 中国棉花, 2009, 36(7): 2-3. |
| XU Hongxia, YANG Weihua, WANG Yanqin, et al. Analysis on the quality of oil cottonseed in China[J]. China Cotton, 2009, 36(7): 2-3. | |
| [2] |
Liu Q, Singh S, Green A. Genetic modification of cotton seed oil using inverted-repeat gene-silencing techniques[J]. Biochemical Society Transactions, 2000, 28(6): 927-929.
PMID |
| [3] | Meneghetti S M P, Meneghetti M R, Serra T M, et al. Biodiesel production from vegetable oil mixtures: cottonseed, soybean, and Castor oils[J]. Energy & Fuels, 2007, 21(6): 3746-3747. |
| [4] | 贾秀凌. 陆地棉SSR标记遗传图谱构建与纤维品质性状QTL定位[D]. 重庆: 西南大学, 2014. |
| JIA Xiuling. Construction of upland cotton SSR marker genetic map and QTL mapping of fiber quality traits[D]. Chongqing: Southwest University, 2014. | |
| [5] |
Huang X H, Han B. Natural variations and genome-wide association studies in crop plants[J]. Annual Review of Plant Biology, 2014, 65: 531-551.
DOI PMID |
| [6] |
Yano K, Yamamoto E, Aya K, et al. Genome-wide association study using whole-genome sequencing rapidly identifies new genes influencing agronomic traits in rice[J]. Nature Genetics, 2016, 48(8): 927-934.
DOI PMID |
| [7] | Yang N, Lu Y L, Yang X H, et al. Genome wide association studies using a new nonparametric model reveal the genetic architecture of 17 agronomic traits in an enlarged maize association panel[J]. PLoS Genetics, 2014, 10(9): e1004573. |
| [8] |
Han Y P, Zhao X, Liu D Y, et al. Domestication footprints anchor genomic regions of agronomic importance in soybeans[J]. New Phytologist, 2016, 209(2): 871-884.
DOI PMID |
| [9] | 刘凯, 邓志英, 张莹, 等. 小麦茎秆断裂强度相关性状QTL的连锁和关联分析[J]. 作物学报, 2017, 43(4): 483-495. |
| LIU Kai, DENG Zhiying, ZHANG Ying, et al. Linkage analysis and genome-wide association study of QTLs controlling stem-breaking-strength-related traits in wheat[J]. Acta Agronomica Sinica, 2017, 43(4): 483-495. | |
| [10] | Yuan Y C, Wang X L, Wang L Y, et al. Genome-wide association study identifies candidate genes related to seed oil composition and protein content in Gossypium hirsutum L[J]. Frontiers in Plant Science, 2018, 9: 1359. |
| [11] | Zhao W X, Kong X H, Yang Y, et al. Association mapping seed kernel oil content in upland cotton using genome-wide SSRs and SNPs[J]. Molecular Breeding, 2019, 39(7): 105. |
| [12] | Liu D X, Liu F, Shan X R, et al. Construction of a high-density genetic map and lint percentage and cottonseed nutrient trait QTL identification in upland cotton (Gossypium hirsutum L.)[J]. Molecular Genetics and Genomics, 2015, 290(5): 1683-1700. |
| [13] |
Li X M, Yuan D J, Wang H T, et al. Increasing cotton genome coverage with polymorphic SSRs as revealed by SSCP[J]. Genome, 2012, 55(6): 459-470.
DOI PMID |
| [14] |
Nie X H, Huang C, You C Y, et al. Genome-wide SSR-based association mapping for fiber quality in nation-wide upland cotton inbreed cultivars in China[J]. BMC Genomics, 2016, 17: 352.
DOI PMID |
| [15] |
田博宇, 黄义文, 周大云, 等. 不同形态棉籽中油分含量快速无损近红外检测方法的建立[J]. 棉花学报, 2023, 35(4): 325-333.
DOI |
| TIAN Boyu, HUANG Yiwen, ZHOU Dayun, et al. Development of a rapid non-destructive method for the detection of cottonseed oil content by near-infrared spectroscopy[J]. Cotton Science, 2023, 35(4): 325-333. | |
| [16] |
Evanno G, Regnaut S, Goudet J. Detecting the number of clusters of individuals using the software STRUCTURE: a simulation study[J]. Molecular Ecology, 2005, 14(8): 2611-2620.
DOI PMID |
| [17] |
Falush D, Stephens M, Pritchard J K. Inference of population structure using multilocus genotype data: dominant markers and null alleles[J]. Molecular Ecology Notes, 2007, 7(4): 574-578.
DOI PMID |
| [18] |
Liu K J, Muse S V. PowerMarker: an integrated analysis environment for genetic marker analysis[J]. Bioinformatics, 2005, 21(9): 2128-2129.
DOI PMID |
| [19] |
雷杰杰, 邵盘霞, 郭春平, 等. 新疆陆地棉经济性状优异等位基因位点的遗传解析[J]. 棉花学报, 2020, 32(3): 185-198.
DOI |
| LEI Jiejie, SHAO Panxia, GUO Chunping, et al. Genetic dissection of allelic loci associated with economic traits of upland cottons in Xinjiang[J]. Cotton Science, 2020, 32(3): 185-198. | |
| [20] |
Yu Y, Yuan D J, Liang S G, et al. Genome structure of cotton revealed by a genome-wide SSR genetic map constructed from a BC1 population between Gossypium hirsutum and G. barbadense[J]. BMC Genomics, 2011, 12: 15.
DOI PMID |
| [21] | Guo W Z, Cai C P, Wang C B, et al. A microsatellite-based, gene-rich linkage map reveals genome structure, function and evolution in Gossypium[J]. Genetics, 2007, 176(1): 527-541. |
| [22] | Yu J W, Zhang K, Li S Y, et al. Mapping quantitative trait loci for lint yield and fiber quality across environments in a Gossypium hirsutum × Gossypium barbadense backcross inbred line population[J]. Theoretical and Applied Genetics, 2013, 126(1): 275-287. |
| [23] | Wang H T, Huang C, Guo H L, et al. QTL mapping for fiber and yield traits in upland cotton under multiple environments[J]. PLoS One, 2015, 10(6): e0130742. |
| [24] | 王寒涛. 陆地棉遗传图谱的构建及其重要农艺性状的QTL定位[D]. 武汉: 华中农业大学, 2015. |
| WANG Hantao. Construction of upland cotton genetic map and QTL positioning of important agronomic traits[D]. Wuhan: Huazhong Agricultural University, 2015. | |
| [25] |
Du X M, Huang G, He S P, et al. Resequencing of 243 diploid cotton accessions based on an updated A genome identifies the genetic basis of key agronomic traits[J]. Nature Genetics, 2018, 50(6): 796-802.
DOI PMID |
| [26] | Guo W Z, Ma G J, Zhu Y C, et al. Molecular tagging and mapping of quantitative trait loci for lint percentage and morphological marker genes in upland cotton[J]. Journal of Integrative Plant Biology, 2006, 48(3): 320-326. |
| [27] | Sun F D, Zhang J H, Wang S F, et al. QTL mapping for fiber quality traits across multiple generations and environments in upland cotton[J]. Molecular Breeding, 2012, 30(1): 569-582. |
| [28] | Yu J W, Yu S X, Gore M, et al. Identification of quantitative trait loci across interspecific F2, F2: 3 and testcross populations for agronomic and fiber traits in tetraploid cotton[J]. Euphytica, 2013, 191(3): 375-389. |
| [1] | 罗影, 努尔孜亚·亚力买买提, 贾文捷, 朱琦, 石文婷, 李雨桐, 贾培松. 新疆野生平菇的遗传多样性分析[J]. 新疆农业科学, 2025, 62(5): 1266-1272. |
| [2] | 王帆, 李玉姗, 王威, 邓超宏, 赵连佳, 马越, 肖菁, 庄红梅, 许红军. 基于表型性状鉴定及SSR分子标记分析芜菁种质资源遗传多样性[J]. 新疆农业科学, 2024, 61(11): 2601-2613. |
| [3] | 李超, 杨英, 郑贺云, 杨建丽, 陈伟, 杨咪, 孙玉萍. 基于SSR荧光标记分析新疆甜瓜种质资源遗传多样性与群体结构[J]. 新疆农业科学, 2024, 61(11): 2614-2625. |
| [4] | 张雁飞, 苏比娜·肖克来提, 杨磊, 郝庆, 靳娟, 樊丁宇. 26个鲜食枣品种SSR指纹图谱构建与遗传多样性分析[J]. 新疆农业科学, 2023, 60(7): 1671-1678. |
| [5] | 文佳, 黄陈珏, 嵇子涵, 李黎贝, 冯震, 喻树迅. 陆地棉动态株高与SSR标记的关联分析[J]. 新疆农业科学, 2023, 60(12): 2892-2901. |
| [6] | 罗影, 努尔孜亚·亚力买买提, 贾文捷, 刘洁莹, 贾培松. 新疆野生大环柄菇的分子鉴定与遗传多样性分析[J]. 新疆农业科学, 2023, 60(10): 2501-2508. |
| [7] | 马君, 杨延龙, 师维军, 汪鹏龙, 郑巨云, 郭仁松, 胡文冉, 杨洋. 棉花陆海渐渗杂交群体纤维品质性状的QTL定位[J]. 新疆农业科学, 2023, 60(1): 11-16. |
| [8] | 梁雎, 刘国宏, 王保民, 何桥, 阿布来克·尼亚孜, 郭红梅, 李兴婷, 任红松. 不同来源葡萄种质资源果实叶酸含量及ISSR比较分析[J]. 新疆农业科学, 2023, 60(1): 116-126. |
| [9] | 周勃, 任海龙, 张龑, 高强, 徐麟, 邹集文. 金花菜与苜蓿属主要物种基因组SSR分布特征的比较分析[J]. 新疆农业科学, 2022, 59(9): 2217-2223. |
| [10] | 吴巧玉, 何天久. 紫色甘薯资源的形态学和ISSR荧光标记分析[J]. 新疆农业科学, 2022, 59(7): 1625-1631. |
| [11] | 巴爱丽, 杨靖, 贾菲芸, 樊苗苗, 张冉, 李友勇. 玉米杂种优势类群划分高多态SSR引物筛选[J]. 新疆农业科学, 2022, 59(6): 1373-1383. |
| [12] | 陆小双, 张梦洁, 韩万里, 龙遗磊, 刘鹏飞, 陈全家, 曲延英, 邓晓娟. 海岛棉抗枯萎病性状与 SSR标记的关联分析[J]. 新疆农业科学, 2022, 59(10): 2365-2373. |
| [13] | 马光皇, 康淑媛, 李莉, 肖海兵, 熊仁次, 杨明禄. 枣瘿蚊微卫星标记筛选及微卫星多态性分析[J]. 新疆农业科学, 2020, 57(5): 888-894. |
| [14] | 李寐华, 杨永, 马新力, 张学军, 张红, 张永兵, 伊鸿平. 基于SSR分子标记对西瓜杂交种早佳8424纯度的高通量鉴定[J]. 新疆农业科学, 2020, 57(3): 464-469. |
| [15] | 王勇, 孙锋, 苏来曼·艾则孜, 李玉玲, 伍国红, 骆强伟, 郭平峰. 19个自育葡萄品种(系)指纹图谱构建[J]. 新疆农业科学, 2020, 57(12): 2238-2249. |
| 阅读次数 | ||||||
|
全文 |
|
|||||
|
摘要 |
|
|||||