

新疆农业科学 ›› 2025, Vol. 62 ›› Issue (1): 182-192.DOI: 10.6048/j.issn.1001-4330.2025.01.021
范蓉1( ), 张永兵1, 李寐华1, 张学军1, 伊鸿平1, 刘钊2, 杨永1(
), 张永兵1, 李寐华1, 张学军1, 伊鸿平1, 刘钊2, 杨永1( )
)
收稿日期:2024-07-17
出版日期:2025-01-20
发布日期:2025-03-11
通信作者:
杨永(1986-),男,山东曹县人,副研究员,硕士,研究方向为甜瓜遗传分子改良,(E-mail)553508458@qq.com作者简介:范蓉(1995-),女,新疆塔城人,助理研究员,硕士,研究方向为甜瓜遗传分子改良,(E-mail)1029681312@qq.com
基金资助:
FAN Rong1( ), ZHANG Yongbing1, LI Meihua1, ZHANG Xuejun1, YI Hongping1, LIU Zhao2, YANG Yong1(
), ZHANG Yongbing1, LI Meihua1, ZHANG Xuejun1, YI Hongping1, LIU Zhao2, YANG Yong1( )
)
Received:2024-07-17
Published:2025-01-20
Online:2025-03-11
Supported by:摘要:
【目的】 研究厚皮甜瓜果实心部可溶性固形物的遗传规律和QTL定位,挖掘厚皮甜瓜果实心部可溶性固形物相关的候选基因。【方法】 以高糖材料P1和低糖材料P2为双亲构建六世代分离群体;采用主基因+多基因混合遗传模型研究厚皮甜瓜心部果肉可溶性固形物含量的遗传规律,并基于F2群体,选取果实可溶性固形物含量极端的单株构建混池,利用BSA方法对甜瓜可溶性固形物含量进行定位。【结果】 甜瓜可溶性固形物符合E-1(MX2-ADI-AD)遗传模型,2对主基因以上位性效应为主,其次为显性效应、加性效应。2个QTL分别在第5号染色体827066 bp-109953 bp和第8号11316600 bp-11729324 bp,区间大小分别为0.27 Mb和0.41 Mb,2个区间内共包含50个候选基因。筛选获得了6个与甜瓜可溶性固形物含量相关的候选基因,分别是MELO3C014619、MELO3C014617、MELO3C014596、MELO3C014594、MELO3C019077、MELO3C019089。【结论】 厚皮甜瓜心部可溶性固形物符合E-1(MX2-ADI-AD)遗传模型,通过BSA方法在5号和8号染色体定位到2个甜瓜可溶性固形物相关QTL,筛选出6个与甜瓜可溶性固形物含量相关的候选基因。
中图分类号:
范蓉, 张永兵, 李寐华, 张学军, 伊鸿平, 刘钊, 杨永. 厚皮甜瓜心部果肉可溶性固形物含量遗传规律分析及QTL定位[J]. 新疆农业科学, 2025, 62(1): 182-192.
FAN Rong, ZHANG Yongbing, LI Meihua, ZHANG Xuejun, YI Hongping, LIU Zhao, YANG Yong. QTL mapping and genetic analysis of soluble solids content in the center flesh of muskmelon[J]. Xinjiang Agricultural Sciences, 2025, 62(1): 182-192.
| 世代 Generations | 次数分布Frequency distribution | 平均值 Average(°Brix) | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 4 | 5 | 6 | 7 | 8 | 9 | 10 | 11 | 12 | 13 | 14 | 15 | ||
| P1 | 2 | 6 | 11 | 8 | 14.49±0.89aA | ||||||||
| P2 | 3 | 7 | 5 | 2 | 3 | 7.34±1.23dD | |||||||
| F1 | 1 | 2 | 1 | 2 | 10 | 17 | 4 | 2 | 11.93±1.46aA | ||||
| B1 | 2 | 1 | 1 | 7 | 13 | 24 | 9 | 13.68±1.54bB | |||||
| B2 | 1 | 3 | 4 | 5 | 5 | 11 | 14 | 11 | 4 | 9.48±2.02cC | |||
| F2 | 4 | 7 | 21 | 21 | 30 | 35 | 60 | 101 | 79 | 36 | 4 | 11.69±2.10bB | |
表1 厚皮甜瓜6个群体心部可溶性固形物的次数分布及平均值.
Tab.1 Frequency distribution and average of non-segregating generations of muskmelon heart suger content traits
| 世代 Generations | 次数分布Frequency distribution | 平均值 Average(°Brix) | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 4 | 5 | 6 | 7 | 8 | 9 | 10 | 11 | 12 | 13 | 14 | 15 | ||
| P1 | 2 | 6 | 11 | 8 | 14.49±0.89aA | ||||||||
| P2 | 3 | 7 | 5 | 2 | 3 | 7.34±1.23dD | |||||||
| F1 | 1 | 2 | 1 | 2 | 10 | 17 | 4 | 2 | 11.93±1.46aA | ||||
| B1 | 2 | 1 | 1 | 7 | 13 | 24 | 9 | 13.68±1.54bB | |||||
| B2 | 1 | 3 | 4 | 5 | 5 | 11 | 14 | 11 | 4 | 9.48±2.02cC | |||
| F2 | 4 | 7 | 21 | 21 | 30 | 35 | 60 | 101 | 79 | 36 | 4 | 11.69±2.10bB | |
| 模型代号 Model code | 模型 Model | 极大似然函数值 Max-likelihood value | AIC值 AIC value |
|---|---|---|---|
| A-1 | 1MG-AD | -1 215.89 | 2 439.77 |
| A-2 | 1MG-A | -1 251.65 | 2 509.29 |
| A-3 | 1MG-EAD | -1 250.56 | 2 507.13 |
| A-4 | 1MG-NCD | -1 341.19 | 2 688.38 |
| B-1 | 2MG-ADI | -1 188.81 | 2 397.62 |
| B-2 | 2MG-AD | -1 206.98 | 2 425.95 |
| B-3 | 2MG-A | -1 277.68 | 2 563.36 |
| B-4 | 2MG-EA | -1 225.30 | 2 456.59 |
| B-5 | 2MG-CD | -1 232.31 | 2 472.61 |
| B-6 | 2MG-EAD | -1 233.54 | 2 473.07 |
| C-0 | PG-ADI | -1 226.94 | 2 473.89 |
| C-1 | PG-AD | -1 229.72 | 2 473.43 |
| D-0 | MX1-AD-ADI | -1 173.92 | 2 371.84 |
| D-1 | MX1-AD-AD | -1 229.08 | 2 476.15 |
| D-2 | MX1-A-AD | -1 229.68 | 2 475.35 |
| D-3 | MX1-EAD-AD | -1 187.83 | 2 391.66 |
| D-4 | MX1-NCD-AD | -1 229.70 | 2 475.41 |
| E-0 | MX2-ADI-ADI | -1 172.85 | 2 381.69 |
| E-1 | MX2-ADI-AD | -1 169.32 | 2 368.64 |
| E-2 | MX2-AD-AD | -1 229.70 | 2 481.40 |
| E-3 | MX2-A-AD | -1 194.86 | 2 407.71 |
| E-4 | MX2-EA-AD | -1 229.66 | 2 475.33 |
| E-5 | MX2-CD-AD | -1 187.59 | 2 393.17 |
| E-6 | MX2-EAD-AD | -1 229.69 | 2 475.38 |
表2 不同遗传模型的极大似然值和AIC值
Tab.2 Max-likelihood-value and AIC value of the different genetic models
| 模型代号 Model code | 模型 Model | 极大似然函数值 Max-likelihood value | AIC值 AIC value |
|---|---|---|---|
| A-1 | 1MG-AD | -1 215.89 | 2 439.77 |
| A-2 | 1MG-A | -1 251.65 | 2 509.29 |
| A-3 | 1MG-EAD | -1 250.56 | 2 507.13 |
| A-4 | 1MG-NCD | -1 341.19 | 2 688.38 |
| B-1 | 2MG-ADI | -1 188.81 | 2 397.62 |
| B-2 | 2MG-AD | -1 206.98 | 2 425.95 |
| B-3 | 2MG-A | -1 277.68 | 2 563.36 |
| B-4 | 2MG-EA | -1 225.30 | 2 456.59 |
| B-5 | 2MG-CD | -1 232.31 | 2 472.61 |
| B-6 | 2MG-EAD | -1 233.54 | 2 473.07 |
| C-0 | PG-ADI | -1 226.94 | 2 473.89 |
| C-1 | PG-AD | -1 229.72 | 2 473.43 |
| D-0 | MX1-AD-ADI | -1 173.92 | 2 371.84 |
| D-1 | MX1-AD-AD | -1 229.08 | 2 476.15 |
| D-2 | MX1-A-AD | -1 229.68 | 2 475.35 |
| D-3 | MX1-EAD-AD | -1 187.83 | 2 391.66 |
| D-4 | MX1-NCD-AD | -1 229.70 | 2 475.41 |
| E-0 | MX2-ADI-ADI | -1 172.85 | 2 381.69 |
| E-1 | MX2-ADI-AD | -1 169.32 | 2 368.64 |
| E-2 | MX2-AD-AD | -1 229.70 | 2 481.40 |
| E-3 | MX2-A-AD | -1 194.86 | 2 407.71 |
| E-4 | MX2-EA-AD | -1 229.66 | 2 475.33 |
| E-5 | MX2-CD-AD | -1 187.59 | 2 393.17 |
| E-6 | MX2-EAD-AD | -1 229.69 | 2 475.38 |
| 模型 Model | 世代 Generation | 拟合模型的统计参数Statistic parameter of fifit model | ||||
|---|---|---|---|---|---|---|
| nW2 | Dn | |||||
| D-0 MX1-AD-ADI | P1 | 0.015 6(0.900 6) | 0.032 6(0.856 7) | 0.056 7(0.811 7) | 0.042 2(0.921 4) | 0.104 4(0.900 8) |
| F1 | 0.300 5(0.583 6) | 0.017 5(0.894 7) | 2.539 3(0.111 0) | 0.243 8(0.201 6) | 0.193 4(0.094 3) | |
| P2 | 0.073 8(0.785 8) | 0.044 5(0.833 0) | 0.043 6(0.834 6) | 0.070 9(0.753 4) | 0.123 1(0.886 8) | |
| B1 | 1.028 6(0.310 5) | 0.114 9(0.734 7) | 6.616 5(0.010 1)* | 0.481 1(0.045 5)* | 0.162(0.089 5) | |
| B2 | 0.022 5(0.880 8) | 0.147 3(0.701 1) | 0.911 3(0.339 8) | 0.044 3(0.909 7) | 0.068 1(0.933 9) | |
| F2 | 0.155 3(0.693 5) | 0.108 0(0.742 4) | 0.044 8(0.832 3) | 0.097 8(0.607 2) | 0.045 9(0.372 5) | |
| E-0 MX2-ADI-ADI | P1 | 0.015 6(0.900 5) | 0.032 6(0.856 7) | 0.056 6(0.812 0) | 0.042 2(0.921 3) | 0.104 3(0.901 1) |
| F1 | 0.300 2(0.583 8) | 0.017 5(0.894 9) | 2.539 4(0.111 0) | 0.243 8(0.201 7) | 0.193 3(0.094 4) | |
| P2 | 0.073 8(0.785 9) | 0.044 4(0.833 1) | 0.043 7(0.834 4) | 0.070 9(0.753 5) | 0.123 0(0.887 0) | |
| B1 | 1.030 0(0.310 2) | 0.126 8(0.721 8) | 6.282 1(0.012 2)* | 0.469 5(0.048 8)* | 0.161 4(0.091 7) | |
| B2 | 0.003 2(0.954 7) | 0.032 2(0.857 6) | 0.879 2(0.348 4) | 0.069 6(0.760 5) | 0.097 2(0.609 1) | |
| F2 | 0.058 9(0.808 2) | 0.047 5(0.827 5) | 0.004 7(0.945 4) | 0.075 8(0.725 7) | 0.040 9(0.519 9) | |
| E-1 MX2-ADI-AD | P1 | 0.844 6(0.358 1) | 1.001 4(0.317 0) | 0.196 7(0.657 4) | 0.135 4(0.440 5) | 0.167 7(0.390 1) |
| F1 | 0.685 0(0.407 9) | 0.185 5(0.666 7) | 2.198 9(0.138 1) | 0.290 4(0.150 7) | 0.209 7(0.055 5) | |
| P2 | 0.275 9(0.599 4) | 0.183 2(0.668 6) | 0.103 8(0.747 3) | 0.099 6(0.597 6) | 0.145 3(0.739 3) | |
| B1 | 0.373 7(0.541 0) | 1.802 1(0.179 5) | 9.012(0.002 7)* | 0.329 5(0.117 7) | 0.155 8(0.112 6) | |
| B2 | 0.003 6(0.951 8) | 0.100 6(0.751 1) | 1.070 3(0.300 9) | 0.051 8(0.866 1) | 0.079 8(0.825 6) | |
| F2 | 0.213 2(0.644 3) | 0.284 9(0.593 5) | 0.120 3(0.728 7) | 0.095 1(0.621 0) | 0.048 8(0.299 0) | |
表3 不同世代果实心糖性状拟合优度模型检验
Tab.3 Tests for goodness of fifit model of heart suger traits in different generations
| 模型 Model | 世代 Generation | 拟合模型的统计参数Statistic parameter of fifit model | ||||
|---|---|---|---|---|---|---|
| nW2 | Dn | |||||
| D-0 MX1-AD-ADI | P1 | 0.015 6(0.900 6) | 0.032 6(0.856 7) | 0.056 7(0.811 7) | 0.042 2(0.921 4) | 0.104 4(0.900 8) |
| F1 | 0.300 5(0.583 6) | 0.017 5(0.894 7) | 2.539 3(0.111 0) | 0.243 8(0.201 6) | 0.193 4(0.094 3) | |
| P2 | 0.073 8(0.785 8) | 0.044 5(0.833 0) | 0.043 6(0.834 6) | 0.070 9(0.753 4) | 0.123 1(0.886 8) | |
| B1 | 1.028 6(0.310 5) | 0.114 9(0.734 7) | 6.616 5(0.010 1)* | 0.481 1(0.045 5)* | 0.162(0.089 5) | |
| B2 | 0.022 5(0.880 8) | 0.147 3(0.701 1) | 0.911 3(0.339 8) | 0.044 3(0.909 7) | 0.068 1(0.933 9) | |
| F2 | 0.155 3(0.693 5) | 0.108 0(0.742 4) | 0.044 8(0.832 3) | 0.097 8(0.607 2) | 0.045 9(0.372 5) | |
| E-0 MX2-ADI-ADI | P1 | 0.015 6(0.900 5) | 0.032 6(0.856 7) | 0.056 6(0.812 0) | 0.042 2(0.921 3) | 0.104 3(0.901 1) |
| F1 | 0.300 2(0.583 8) | 0.017 5(0.894 9) | 2.539 4(0.111 0) | 0.243 8(0.201 7) | 0.193 3(0.094 4) | |
| P2 | 0.073 8(0.785 9) | 0.044 4(0.833 1) | 0.043 7(0.834 4) | 0.070 9(0.753 5) | 0.123 0(0.887 0) | |
| B1 | 1.030 0(0.310 2) | 0.126 8(0.721 8) | 6.282 1(0.012 2)* | 0.469 5(0.048 8)* | 0.161 4(0.091 7) | |
| B2 | 0.003 2(0.954 7) | 0.032 2(0.857 6) | 0.879 2(0.348 4) | 0.069 6(0.760 5) | 0.097 2(0.609 1) | |
| F2 | 0.058 9(0.808 2) | 0.047 5(0.827 5) | 0.004 7(0.945 4) | 0.075 8(0.725 7) | 0.040 9(0.519 9) | |
| E-1 MX2-ADI-AD | P1 | 0.844 6(0.358 1) | 1.001 4(0.317 0) | 0.196 7(0.657 4) | 0.135 4(0.440 5) | 0.167 7(0.390 1) |
| F1 | 0.685 0(0.407 9) | 0.185 5(0.666 7) | 2.198 9(0.138 1) | 0.290 4(0.150 7) | 0.209 7(0.055 5) | |
| P2 | 0.275 9(0.599 4) | 0.183 2(0.668 6) | 0.103 8(0.747 3) | 0.099 6(0.597 6) | 0.145 3(0.739 3) | |
| B1 | 0.373 7(0.541 0) | 1.802 1(0.179 5) | 9.012(0.002 7)* | 0.329 5(0.117 7) | 0.155 8(0.112 6) | |
| B2 | 0.003 6(0.951 8) | 0.100 6(0.751 1) | 1.070 3(0.300 9) | 0.051 8(0.866 1) | 0.079 8(0.825 6) | |
| F2 | 0.213 2(0.644 3) | 0.284 9(0.593 5) | 0.120 3(0.728 7) | 0.095 1(0.621 0) | 0.048 8(0.299 0) | |
| 一阶遗传参数 1st order parameter | 估值Estimate | 二阶遗传参数 2nd order parameter | 估值Estimate | ||
|---|---|---|---|---|---|
| E-1 | B1 | B2 | F2 | ||
| m | 12.4107 | 0.952 1 | 2.827 5 | 3.167 7 | |
| da | -0.050 7 | 0.19 | 0 | 0 | |
| db | -0.050 7 | 39.956 9 | 69.502 1 | 71.855 4 | |
| ha | 0.953 2 | 7.974 8 | 0.000 2 | 0.000 2 | |
| hb | -1.419 | ||||
| i | -1.532 9 | ||||
| jab | 2.358 4 | ||||
| jba | -0.013 8 | ||||
| l | 1.571 | ||||
| [d] | 3.571 5 | ||||
| [h] | -1.651 8 | ||||
表4 甜瓜心糖性状拟合模型的遗传参数估值
Tab.4 The estimation of genetic parameters of fifit model of melon heart suger resistance traits
| 一阶遗传参数 1st order parameter | 估值Estimate | 二阶遗传参数 2nd order parameter | 估值Estimate | ||
|---|---|---|---|---|---|
| E-1 | B1 | B2 | F2 | ||
| m | 12.4107 | 0.952 1 | 2.827 5 | 3.167 7 | |
| da | -0.050 7 | 0.19 | 0 | 0 | |
| db | -0.050 7 | 39.956 9 | 69.502 1 | 71.855 4 | |
| ha | 0.953 2 | 7.974 8 | 0.000 2 | 0.000 2 | |
| hb | -1.419 | ||||
| i | -1.532 9 | ||||
| jab | 2.358 4 | ||||
| jba | -0.013 8 | ||||
| l | 1.571 | ||||
| [d] | 3.571 5 | ||||
| [h] | -1.651 8 | ||||
| 样本 Sample | 原始Reads Raw reads | 原始数据量 Raw bases | 过滤后Reads Clean reads | 过滤后 碱基数 Clean bases | 有效Reads比 Valid reads (%) | 有效数 据量比 Valid bases (%) | GC (%) | Q20 (%) | Q30 (%) |
|---|---|---|---|---|---|---|---|---|---|
| HS | 38 148 520 | 11 145 445 500 | 75 572 218 | 11 045 883 000 | 99.05 | 99.11 | 36.72 | 95.56 | 89.04 |
| LS | 37 150 247 | 11 444 585 400 | 73 639 220 | 11 335 832 700 | 99.11 | 99.05 | 37.86 | 95.56 | 89.1 |
| P1 | 27 381 098 | 8 214 073 200 | 54 493 862 | 8 174 079 300 | 99.51 | 99.51 | 36.39 | 95.68 | 89.12 |
| P2 | 27 496 523 | 8 248 618 800 | 54 723 580 | 8 208 537 000 | 99.51 | 99.51 | 37.23 | 95.64 | 89.13 |
表5 原始数据统计
Tab.5 Quality statistics of raw data
| 样本 Sample | 原始Reads Raw reads | 原始数据量 Raw bases | 过滤后Reads Clean reads | 过滤后 碱基数 Clean bases | 有效Reads比 Valid reads (%) | 有效数 据量比 Valid bases (%) | GC (%) | Q20 (%) | Q30 (%) |
|---|---|---|---|---|---|---|---|---|---|
| HS | 38 148 520 | 11 145 445 500 | 75 572 218 | 11 045 883 000 | 99.05 | 99.11 | 36.72 | 95.56 | 89.04 |
| LS | 37 150 247 | 11 444 585 400 | 73 639 220 | 11 335 832 700 | 99.11 | 99.05 | 37.86 | 95.56 | 89.1 |
| P1 | 27 381 098 | 8 214 073 200 | 54 493 862 | 8 174 079 300 | 99.51 | 99.51 | 36.39 | 95.68 | 89.12 |
| P2 | 27 496 523 | 8 248 618 800 | 54 723 580 | 8 208 537 000 | 99.51 | 99.51 | 37.23 | 95.64 | 89.13 |
| 样本 Sample | 过滤后Reads Clean reads | 比对上数据 Mapping reads | 比对率 Mapped (%) | 平均测序深度 Mean coverage | 1×覆盖度 Coverage≥ 1× (%) | 4×覆盖度 Coverage≥ 4× (%) |
|---|---|---|---|---|---|---|
| HS | 75 572 218 | 70 862 228 | 93.77 | 24.55 | 97.8 | 96.74 |
| LS | 73 639 220 | 69 954 351 | 95 | 25.36 | 97.77 | 96.67 |
| P1 | 54 493 862 | 52 222 873 | 95.83 | 19.52 | 96.71 | 95.27 |
| P2 | 54 723 580 | 52 620 862 | 96.16 | 19.2 | 96.86 | 95.42 |
表6 与参考基因组比对结果统计
Tab.6 Matching of quality control data with reference genome
| 样本 Sample | 过滤后Reads Clean reads | 比对上数据 Mapping reads | 比对率 Mapped (%) | 平均测序深度 Mean coverage | 1×覆盖度 Coverage≥ 1× (%) | 4×覆盖度 Coverage≥ 4× (%) |
|---|---|---|---|---|---|---|
| HS | 75 572 218 | 70 862 228 | 93.77 | 24.55 | 97.8 | 96.74 |
| LS | 73 639 220 | 69 954 351 | 95 | 25.36 | 97.77 | 96.67 |
| P1 | 54 493 862 | 52 222 873 | 95.83 | 19.52 | 96.71 | 95.27 |
| P2 | 54 723 580 | 52 620 862 | 96.16 | 19.2 | 96.86 | 95.42 |
| 染色体编号 Chromosome ID | 方法 Methods | 关联区间起始位置 Start position of the correlation interval(bp) | 关联区间终止位置 End position of the correlation interval(bp) | 关联区间大小 Correlation interval size (Mb) |
|---|---|---|---|---|
| Chr5 | Deep Learning | 805192 | 1196476 | 0.391284 |
| K value | 660399 | 1099530 | 0.439131 | |
| ED4 | 684566 | 1099550 | 0.414984 | |
| △SNP index | 740823 | 1187084 | 0.446261 | |
| SmoothG | 738888 | 1185404 | 0.446516 | |
| SmoothLOD | 827066 | 1196504 | 0.369438 | |
| Ridit | 735951 | 1108157 | 0.372206 | |
| Chr8 | Deep Learning | 11093331 | 11729324 | 0.635993 |
| K value | 11115505 | 11773204 | 0.657699 | |
| ED4 | 11316600 | 14061373 | 2.744773 | |
| △SNP index | 11115505 | 11773204 | 0.657699 | |
| SmoothG | 11115620 | 11779123 | 0.663503 | |
| SmoothLOD | 11316350 | 14061163 | 2.744813 | |
| Ridit | 11164863 | 11808230 | 0.643367 |
表7 7种不同算法的候选区间统计
Tab.7 Statistics of candidate intervals for 7 different algorithms
| 染色体编号 Chromosome ID | 方法 Methods | 关联区间起始位置 Start position of the correlation interval(bp) | 关联区间终止位置 End position of the correlation interval(bp) | 关联区间大小 Correlation interval size (Mb) |
|---|---|---|---|---|
| Chr5 | Deep Learning | 805192 | 1196476 | 0.391284 |
| K value | 660399 | 1099530 | 0.439131 | |
| ED4 | 684566 | 1099550 | 0.414984 | |
| △SNP index | 740823 | 1187084 | 0.446261 | |
| SmoothG | 738888 | 1185404 | 0.446516 | |
| SmoothLOD | 827066 | 1196504 | 0.369438 | |
| Ridit | 735951 | 1108157 | 0.372206 | |
| Chr8 | Deep Learning | 11093331 | 11729324 | 0.635993 |
| K value | 11115505 | 11773204 | 0.657699 | |
| ED4 | 11316600 | 14061373 | 2.744773 | |
| △SNP index | 11115505 | 11773204 | 0.657699 | |
| SmoothG | 11115620 | 11779123 | 0.663503 | |
| SmoothLOD | 11316350 | 14061163 | 2.744813 | |
| Ridit | 11164863 | 11808230 | 0.643367 |
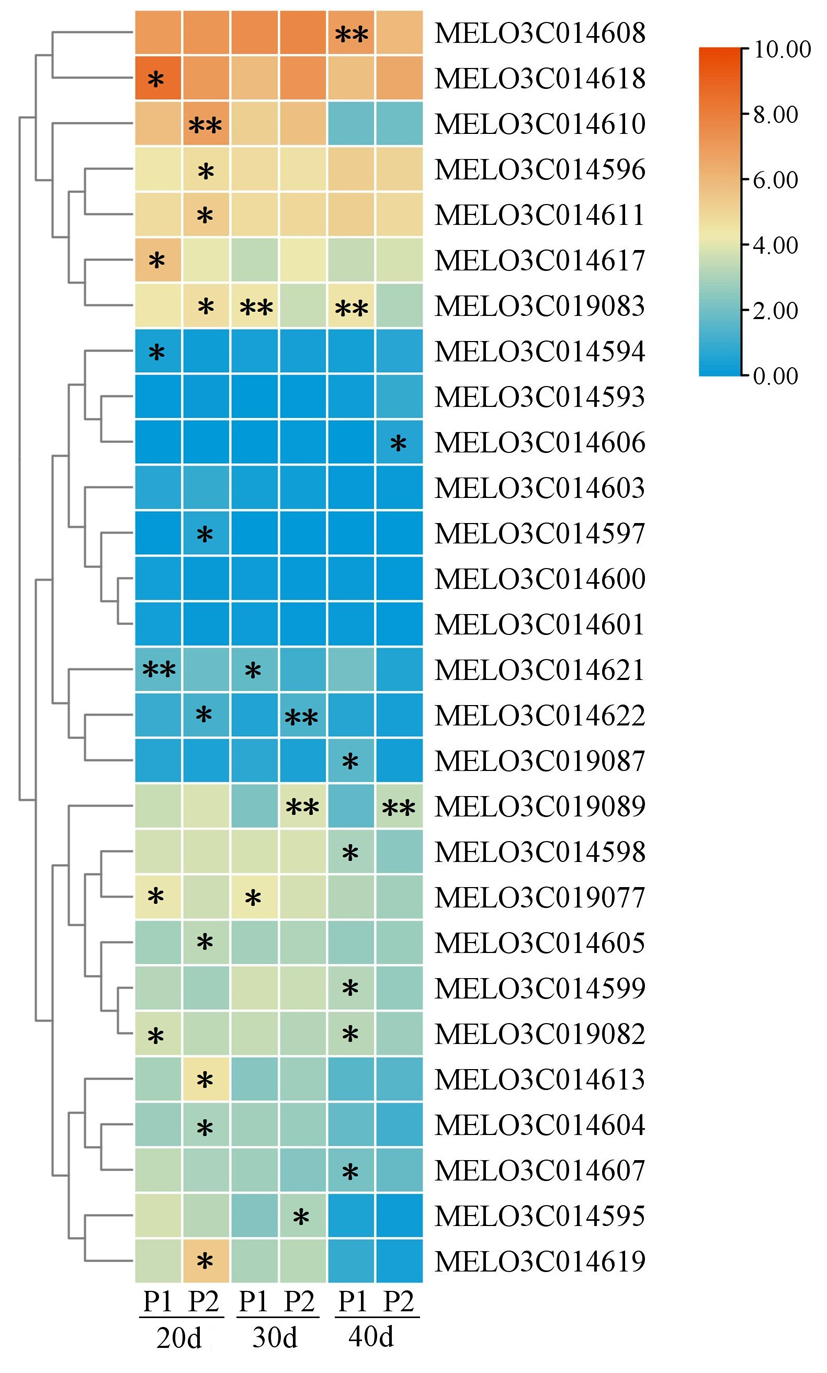
图3 候选基因表达 注:*表示在0.05水平存在显著差异 ; * *表示在0.01水平存在显著差异
Fig.3 Candidate gene expression Notes: *indicate significant differences at the 0.05 leve, * *indicate significant differences at the 0.01 level
| 基因号 Gene ID | 位置 Position | 基因 方向 Gene orientation | 大小 Size (bp) | 注释 Annotation | 编码区非同义突变 Non synonymous mutation sites in coding regions | 启动子区 Promotor Region |
|---|---|---|---|---|---|---|
| MELO3C014622 | Chr5:882794-885290 | - | 2496 | 丝氨酸/苏氨酸蛋白激酶 | 2个SNP | |
| MELO3C014621 | Chr5:889359-892049 | - | 2690 | 含溴结构域的蛋白质 | 1个Indel | |
| MELO3C014619 | Chr5:896637-899328 | - | 2691 | 未知 | 4个SNP | 14个SNP,11个Indel |
| MELO3C014617 | Chr5:905878-908840 | - | 2962 | 未知 | 5个SNP | 29个SNP,7个Indel |
| MELO3C014613 | Chr5:924958-933054 | - | 8096 | DUF1680结构域蛋白 | 6个SNP,6个Indel | |
| MELO3C014610 | Chr5:953260-954563 | - | 1303 | 激酶家族蛋白 | 6个SNP,6个Indel | |
| MELO3C014596 | Chr5:1075795-1079969 | + | 4174 | 苹果酸/酮戊二酸转运蛋白 | 3个SNP | 8个SNP,1个Indel |
| MELO3C014595 | Chr5:1085108-1089566 | + | 4458 | MATE外排家族蛋白 | 10个SNP,1个Indel | |
| MELO3C014594 | Chr5:1091412-1093292 | + | 1880 | 含五肽重复序列的蛋白质 | 4个SNP | 1个Indel |
| MELO3C019077 | Chr8:11363674-11364042 | + | 368 | 液泡融合蛋白CCZ1 | 2个SNP | 22个SNP,5个Indel |
| MELO3C019082 | Chr8:11459977-11463627 | + | 3650 | 血清反应因子结合蛋白1 | 9个SNP,1个Indel | |
| MELO3C019083 | Chr8:11498986-11521642 | + | 22656 | 微管相关蛋白70-2 | 21个SNP,4个Indel | |
| MELO3C019087 | Chr8:11600872-11603741 | + | 2869 | UDP糖基转移酶超家族蛋白 | 18个SNP,7个Indel | |
| MELO3C019089 | Chr8:11687078-11690459 | + | 3381 | DNA促旋酶亚基B | 1个SNP | 21个SNP,2个Indel |
表8 候选基因功能注释
Tab.8 Functional annotation of 3 candidate genes
| 基因号 Gene ID | 位置 Position | 基因 方向 Gene orientation | 大小 Size (bp) | 注释 Annotation | 编码区非同义突变 Non synonymous mutation sites in coding regions | 启动子区 Promotor Region |
|---|---|---|---|---|---|---|
| MELO3C014622 | Chr5:882794-885290 | - | 2496 | 丝氨酸/苏氨酸蛋白激酶 | 2个SNP | |
| MELO3C014621 | Chr5:889359-892049 | - | 2690 | 含溴结构域的蛋白质 | 1个Indel | |
| MELO3C014619 | Chr5:896637-899328 | - | 2691 | 未知 | 4个SNP | 14个SNP,11个Indel |
| MELO3C014617 | Chr5:905878-908840 | - | 2962 | 未知 | 5个SNP | 29个SNP,7个Indel |
| MELO3C014613 | Chr5:924958-933054 | - | 8096 | DUF1680结构域蛋白 | 6个SNP,6个Indel | |
| MELO3C014610 | Chr5:953260-954563 | - | 1303 | 激酶家族蛋白 | 6个SNP,6个Indel | |
| MELO3C014596 | Chr5:1075795-1079969 | + | 4174 | 苹果酸/酮戊二酸转运蛋白 | 3个SNP | 8个SNP,1个Indel |
| MELO3C014595 | Chr5:1085108-1089566 | + | 4458 | MATE外排家族蛋白 | 10个SNP,1个Indel | |
| MELO3C014594 | Chr5:1091412-1093292 | + | 1880 | 含五肽重复序列的蛋白质 | 4个SNP | 1个Indel |
| MELO3C019077 | Chr8:11363674-11364042 | + | 368 | 液泡融合蛋白CCZ1 | 2个SNP | 22个SNP,5个Indel |
| MELO3C019082 | Chr8:11459977-11463627 | + | 3650 | 血清反应因子结合蛋白1 | 9个SNP,1个Indel | |
| MELO3C019083 | Chr8:11498986-11521642 | + | 22656 | 微管相关蛋白70-2 | 21个SNP,4个Indel | |
| MELO3C019087 | Chr8:11600872-11603741 | + | 2869 | UDP糖基转移酶超家族蛋白 | 18个SNP,7个Indel | |
| MELO3C019089 | Chr8:11687078-11690459 | + | 3381 | DNA促旋酶亚基B | 1个SNP | 21个SNP,2个Indel |
| [1] | Eduardo I, Arus P, Monforte A J, et al. Estimating the genetic architecture of fruit quality traits in melon using a genomic library of near isogenic lines[J]. Journal of the American Society for Horticultural Science, 2007, 132(1): 80-89. |
| [2] | Senesi E, Scalzo R L, Prinzivalli C, et al. Relationships between volatile composition and sensory evaluation in eight varieties of netted muskmelon (Cucumis melo L var Reticulatus Naud)[J]. Journal of the Science of Food and Agriculture, 2002, 82(6): 655-662. |
| [3] | Zhang J Z, Li B S, Zhang J L, et al. Organic fertilizer application and Mg fertilizer promote banana yield and quality in an Udic Ferralsol[J]. PLoS One, 2020, 15(3): e0230593. |
| [4] |
Dia M, Wehner T C, Perkins-Veazie P, et al. Stability of fruit quality traits in diverse watermelon cultivars tested in multiple environments[J]. Horticulture Research, 2016, 3: 16066.
DOI PMID |
| [5] | 盖钧镒. 植物数量性状遗传体系[M]. 北京: 科学出版社, 2003. |
| GAI Junyi. Genetic system of quantitative traits in plants[M]. Beijing: Science Press, 2003. | |
| [6] | Qi Z Y, Li J X, Raza M A, et al. Inheritance of fruit cracking resistance of melon (Cucumis melo L.) fitting E-0 genetic model using major gene plus polygene inheritance analysis[J]. Scientia Horticulturae, 2015, 189: 168-174. |
| [7] | Dong J P, Xu J, Xu X W, et al. Inheritance and quantitative trait locus mapping of Fusarium wilt resistance in cucumber[J]. Frontiers in Plant Science, 2019, 10: 1425. |
| [8] | 蒋锋, 闫艳, 黄正刚, 等. 糯玉米种子活力性状的主基因+多基因混合遗传分析[J]. 西北农林科技大学学报(自然科学版), 2023, 51(2): 22-29, 42. |
| JIANG Feng, YAN Yan, HUANG Zhenggang, et al. Mixed major genes and polygenes inheritance analyses for seed vigor traits in waxy corn[J]. Journal of Northwest A & F University (Natural Science Edition), 2023, 51(2): 22-29, 42. | |
| [9] |
解松峰, 吉万全, 张耀元, 等. 小麦重要产量性状的主基因+多基因混合遗传分析[J]. 作物学报, 2020, 46(3): 365-384.
DOI |
|
XIE Songfeng, JI Wanquan, ZHANG Yaoyuan, et al. Genetic effects of important yield traits analysed by mixture model of major gene plus polygene in wheat[J]. Acta Agronomica Sinica, 2020, 46(3): 365-384.
DOI |
|
| [10] | Ren X Y, Li H X, Yin Y, et al. Genetic analysis of fruit traits in wolfberry (Lycium L.) by the major gene plus polygene model[J]. Agronomy, 2022, 12(6): 1403. |
| [11] |
王利民, 党照, 赵玮, 等. 基于分离世代和RIL群体的亚麻株高遗传定位[J]. 植物遗传资源学报, 2022, 23(5): 1446-1457.
DOI |
|
WANG Limin, DANG Zhao, ZHAO Wei, et al. Genetic analysis of plant height in flax using segregating generations and recombination inbred line populations[J]. Journal of Plant Genetic Resources, 2022, 23(5): 1446-1457.
DOI |
|
| [12] | 吕律. 甜瓜果实含糖量遗传分析及糖分积累与蔗糖代谢酶关系的研究[D]. 杭州: 浙江大学, 2018. |
| LYU Lyu. Genetic Analysis of Sugar Content and the Relationship between Sugar Accumulation and Its Metabolism-related Enzymes in Melon(Cucumis melo L.). Hangzhou: Zhejiang University, 2018. | |
| [13] |
许彦宾, 王艳玲, 胡建斌, 等. 甜瓜果实可溶性固形物含量的关联作图及优异等位变异发掘[J]. 园艺学报, 2017, 44(5): 902-910.
DOI |
|
XU Yanbin, WANG Yanling, HU Jianbin, et al. Association mapping of soluble solid content in melon fruits and exploration of the elite alleles[J]. Acta Horticulturae Sinica, 2017, 44(5): 902-910.
DOI |
|
| [14] | Diaz A, Fergany M, Formisano G, et al. A consensus linkage map for molecular markers and quantitative trait loci associated with economically important traits in melon (Cucumis melo L.)[J]. BMC Plant Biology, 2011, 11: 111. |
| [15] | Monforte A J, Oliver M, Gonzalo M J, et al. Identification of quantitative trait loci involved in fruit quality traits in melon (Cucumis melo L.)[J]. TAG Theoretical and Applied Genetics Theoretische und Angewandte Genetik, 2004, 108(4): 750-758. |
| [16] | Paris M K, Zalapa J E, McCreight J D, et al. Genetic dissection of fruit quality components in melon (Cucumis melo L.) using a RIL population derived from exotic × elite US Western Shipping germplasm[J]. Molecular Breeding, 2008, 22(3): 405-419. |
| [17] | Obando J, Fernández-Trujillo J P, Martínez J A, et al. Identification of melon fruit quality quantitative trait loci using near-isogenic lines[J]. Journal of the American Society for Horticultural Science, 2008, 133(1): 139-151. |
| [18] |
王靖天, 张亚雯, 杜应雯, 等. 数量性状主基因+多基因混合遗传分析R软件包SEA v2.0[J]. 作物学报, 2022, 48(6): 1416-1424.
DOI |
|
WANG Jingtian, ZHANG Yawen, DU Yingwen, et al. SEA v2.0: an R software package for mixed major genes plus polygenes inheritance analysis of quantitative traits[J]. Acta Agronomica Sinica, 2022, 48(6): 1416-1424.
DOI |
|
| [19] | Li Z, Chen X X, Shi S Q, et al. DeepBSA: a deep-learning algorithm improves bulked segregant analysis for dissecting complex traits[J]. Molecular Plant, 2022, 15(9): 1418-1427. |
| [20] |
Lingle S E, Dunlap J R. Sucrose metabolism in netted muskmelon fruit during development[J]. Plant Physiology, 1987, 84(2): 386-389.
DOI PMID |
| [21] | 张勇, 马建祥, 李好, 等. 厚皮甜瓜新品种‘农大甜9号’[J]. 园艺学报, 2021, 48(S2):2869-2870. |
|
ZHANG Yong, MA Jianxiang, LI Hao, et al. A new muskmelon cultivar‘nongdatian 9’[J]. Acta Horticulturae Sinica, 2021, 48(S2):2869-2870.
DOI |
|
| [22] | 张若纬, 李肯, 武云鹏, 等. 薄皮甜瓜新品种‘花田1号’[J]. 园艺学报, 2021, 48(S2): 2871-2872. |
|
ZHANG Ruowei, LI Ken, WU Yunpeng, et al. A new oriental melon cultivar‘Huatian 1’[J]. Acta Horticulturae Sinica, 2021, 48(S2): 2871-2872.
DOI |
|
| [23] | Lu J, Qi S Y, Liu R, et al. Nondestructive determination of soluble solids and firmness in mix-cultivar melon using near-infrared CCD spectroscopy[J]. Journal of Innovative Optical Health Sciences, 2015, 8(6): 1550032. |
| [24] | Hu R, Zhang L X, Yu Z Y, et al. Optimization of soluble solids content prediction models in ‘Hami’ melons by means of Vis-NIR spectroscopy and chemometric tools[J]. Infrared Physics & Technology, 2019, 102: 102999. |
| [25] |
郭阳, 史勇, 郭俊先, 等. 近红外光谱技术结合反向区间偏最小二乘算法-连续投影算法预测哈密瓜可溶性固形物含量[J]. 食品与发酵工业, 2022, 48(2): 248-253.
DOI |
|
GUO Yang, SHI Yong, GUO Junxian, et al. Prediction of soluble solids content in Hami melon by combining near-infrared spectroscopy and BiPLS-SPA technology[J]. Food and Fermentation Industries, 2022, 48(2): 248-253.
DOI |
|
| [26] | 叶红霞, 吕律, 海睿, 等. 甜瓜果实糖含量的主基因+多基因遗传分析[J]. 浙江大学学报(农业与生命科学版), 2019, 45(4): 391-400. |
| YE Hongxia, LYU Lyu, HAI Rui, et al. Genetic analysis of fruit sugar content in melon(Cucumis melo L.) based on a mixed model of major genes and polygenes[J]. Journal of Zhejiang University (Agriculture and Life Sciences), 2019, 45(4): 391-400. | |
| [27] | 张宁, 张显, 张勇, 等. 甜瓜远缘群体果实糖含量相关性状遗传分析[J]. 植物遗传资源学报, 2014, 15(5): 932-939. |
|
ZHANG Ning, ZHANG Xian, ZHANG Yong, et al. Genetic analysis of fruit sugar content correlated traits in interspecific population of melon[J]. Journal of Plant Genetic Resources, 2014, 15(5): 932-939.
DOI |
|
| [28] | Pereira L, Ruggieri V, Pérez S, et al. QTL mapping of melon fruit quality traits using a high-density GBS-based genetic map[J]. BMC Plant Biology, 2018, 18(1): 324. |
| [29] |
Argyris J M, Díaz A, Ruggieri V, et al. QTL analyses in multiple populations employed for the fine mapping and identification of candidate genes at a locus affecting sugar accumulation in melon (Cucumis melo L.)[J]. Frontiers in Plant Science, 2017, 8: 1679.
DOI PMID |
| [30] |
Kinoshita H, Nagasaki J, Yoshikawa N, et al. The chloroplastic 2-oxoglutarate/malate transporter has dual function as the malate valve and in carbon/nitrogen metabolism[J]. The Plant Journal, 2011, 65(1): 15-26.
DOI PMID |
| [31] | Schneidereit J, Häusler R E, Fiene G, et al. Antisense repression reveals a crucial role of the plastidic 2-oxoglutarate/malate translocator DiT1 at the interface between carbon and nitrogen metabolism[J]. The Plant Journal, 2006, 45(2): 206-224. |
| [32] |
Zamani-Nour S, Lin H C, Walker B J, et al. Overexpression of the chloroplastic 2-oxoglutarate/malate transporter disturbs carbon and nitrogen homeostasis in rice[J]. Journal of Experimental Botany, 2021, 72(1): 137-152.
DOI PMID |
| [33] |
Li W J, Yang B, Xu J Y, et al. A genome-wide association study reveals that the 2-oxoglutarate/malate translocator mediates seed vigor in rice[J]. The Plant Journal, 2021, 108(2): 478-491.
DOI PMID |
| [34] | Andrade-Marcial M, Pacheco-Arjona R, Góngora-Castillo E, et al. Chloroplastic pentatricopeptide repeat proteins (PPR) in albino plantlets of Agave angustifolia Haw. reveal unexpected behavior[J]. BMC Plant Biology, 2022, 22(1): 352. |
| [35] | Lee K, Park S J, Han J H, et al. A chloroplast-targeted pentatricopeptide repeat protein PPR287 is crucial for chloroplast function and Arabidopsis development[J]. BMC Plant Biology, 2019, 19(1): 244. |
| [36] | Salman M, Sharma P, Kumar M, et al. Targeting novel sites in DNA gyrase for development of anti-microbials[J]. Briefings in Functional Genomics, 2023, 22(2): 180-194. |
| [37] |
Delmas J, Breysse F, Devulder G, et al. Rapid identification of Enterobacteriaceae by sequencing DNA gyrase subunit B encoding gene[J]. Diagnostic Microbiology and Infectious Disease, 2006, 55(4): 263-268.
PMID |
| [38] | Fois B, Skok Ž, TomašiĉT, et al. Dual Escherichia coli DNA gyrase A and B inhibitors with antibacterial activity[J]. ChemMedChem, 2020, 15(3): 265-269. |
| [39] |
Vogt T, Jones P. Glycosyltransferases in plant natural product synthesis: characterization of a supergene family[J]. Trends in Plant Science, 2000, 5(9): 380-386.
DOI PMID |
| [40] | Wei Y Z, Mu H Y, Xu G Z, et al. Genome-wide analysis and functional characterization of the UDP-glycosyltransferase family in grapes[J]. Horticulturae, 2021, 7(8): 204. |
| [41] | Wu B P, Liu X H, Xu K, et al. Genome-wide characterization, evolution and expression profiling of UDP-glycosyltransferase family in pomelo (Citrus grandis) fruit[J]. BMC Plant Biology, 2020, 20(1): 459. |
| [42] |
Wu B P, Cao X M, Liu H R, et al. UDP-glucosyltransferase PpUGT85A2 controls volatile glycosylation in peach[J]. Journal of Experimental Botany, 2019, 70(3): 925-936.
DOI PMID |
| [43] | Liu X G, Lin C L, Ma X D, et al. Functional characterization of a flavonoid glycosyltransferase in sweet orange (Citrus sinensis)[J]. Frontiers in Plant Science, 2018, 9: 166. |
| [1] | 杨君妍, 闫淼, 吴海波, 杨文莉, 王豪杰, 毛建才, 翟文强, 李俊华. 高温对不同厚皮甜瓜品种种子萌发的影响及其耐热性综合评价[J]. 新疆农业科学, 2024, 61(6): 1386-1396. |
| [2] | 沈悦, 凌悦铭, 段晓宇, 杨文莉, 李寐华, 王懿柔, 王惠林, 张学军. 甜瓜抗霜霉病KASP分子标记的开发与验证[J]. 新疆农业科学, 2024, 61(11): 2626-2634. |
| [3] | 贾斌鑫, 杨世英, 王艳, 杨朋朋, 何伟忠, 王成, 刘峰娟, 范盈盈. 不同品种厚皮甜瓜采后糖含量及糖代谢酶活性比较[J]. 新疆农业科学, 2023, 60(6): 1476-1484. |
| [4] | 孙丰磊, 任姣姣, 雷斌, 高文伟, 曲延英. 基于BSA分析技术对陆地棉抗旱基因的鉴定[J]. 新疆农业科学, 2023, 60(10): 2341-2351. |
| [5] | 许铭强, 孟伊娜, 张婷, 马燕, 孟新涛, 邹淑萍, 张谦. 厚皮甜瓜脆片压差膨化干燥工艺[J]. 新疆农业科学, 2022, 59(6): 1429-1437. |
| [6] | 杨永, 范蓉, 张学军, 李寐华, 凌悦铭, 张红, 杨文莉, 姜雪, 张永兵, 伊鸿平. 厚皮甜瓜心部果肉蔗糖含量QTL定位及候选基因分析[J]. 新疆农业科学, 2022, 59(10): 2446-2455. |
| [7] | 袁超, 李晓磊, 孟新涛, 马燕, 徐斌, 张平, 潘俨. 低温贮藏下不同SSC阿克苏富士苹果制汁的适宜性[J]. 新疆农业科学, 2021, 58(4): 607-615. |
| [8] | 凌悦铭, 李寐华, 杨永, 杨文莉, 范蓉, 伊鸿平, 张红, 张学军. 基于BSA-Seq技术的甜瓜抗霜霉病InDel标记开发[J]. 新疆农业科学, 2021, 58(12): 2265-2273. |
| [9] | 杨永, 翟文强, 张学军, 李寐华, 张永兵, 马新力, 伊鸿平. 新疆厚皮甜瓜果实可溶性糖积累规律及其差异分析[J]. 新疆农业科学, 2021, 58(1): 1-8. |
| [10] | 梅闯, 闫鹏, 朱燕飞, 艾沙江·买买提, 马凯, 韩立群, 张振军, 王继勋. 套袋对阿克苏地区富士苹果品质的影响[J]. 新疆农业科学, 2018, 55(9): 1626-1632. |
| [11] | 唐亚萍;杨涛;杨生保;李宁;许娟;帕提古丽;王柏柯;张贵仁;Ann Powell;余庆辉. Heirlooms番茄果实叶绿素和可溶性固形物含量的聚类分析[J]. , 2017, 54(6): 1021-1028. |
| [12] | 田锋;范术丽;魏恒玲;王寒涛;赵树琪;庞朝友;胡守林;喻树迅. 短季棉生育期性状的遗传分析[J]. , 2017, 54(2): 197-205. |
| [13] | 孙玉萍;户金鸽;杨英;张瑞;廖新福. 冬季弱光条件下不同厚皮甜瓜品种生理指标初探[J]. , 2014, 51(9): 1597-1604. |
| [14] | 吴海波;伊鸿平;张永兵;冯炯鑫;郭丽霞. Co60-γ射线对厚皮甜瓜生长及果实性状的影响[J]. , 2014, 51(5): 810-816. |
| [15] | 张学军;季娟;李寐华;王豪杰;伊鸿平. 新疆厚皮甜瓜抗白粉病基因SSR分子标记[J]. , 2014, 51(1): 1-7. |
| 阅读次数 | ||||||
|
全文 |
|
|||||
|
摘要 |
|
|||||