

新疆农业科学 ›› 2024, Vol. 61 ›› Issue (7): 1605-1614.DOI: 10.6048/j.issn.1001-4330.2024.07.006
• 作物遗传育种·种质资源·分子遗传学·耕作栽培·生理生化 • 上一篇 下一篇
杨彩霞1,2( ), 顾炜2,3, 关媛2,3, 瞿静涛2,3, 党冬冬2,3, 吴鹏昊1(
), 顾炜2,3, 关媛2,3, 瞿静涛2,3, 党冬冬2,3, 吴鹏昊1( ), 郑洪建2,3(
), 郑洪建2,3( )
)
收稿日期:2023-12-17
出版日期:2024-07-20
发布日期:2024-09-04
通信作者:
吴鹏昊(1984-),男,新疆人,教授,研究方向为玉米遗传育种,(E-mail)craie788@126.com;作者简介:杨彩霞(1997-),女,重庆人,硕士研究生,研究方向为玉米遗传育种,(E-mail)yangcx4760@163.com
基金资助:
YANG Caixia1,2( ), GU Wei2,3, GUAN Yuan2,3, QU Jingtao2,3, DANG Dongdong2,3, WU Penghao1(
), GU Wei2,3, GUAN Yuan2,3, QU Jingtao2,3, DANG Dongdong2,3, WU Penghao1( ), ZHENG Hongjian2,3(
), ZHENG Hongjian2,3( )
)
Received:2023-12-17
Published:2024-07-20
Online:2024-09-04
Supported by:摘要:
【目的】研究甜玉米基因Sugary1(Su1)序列,分析核苷酸多态性及检验,研究不同来源甜玉米中的Su1的等位变异位点,为甜玉米的品种改良和提高品质提供理论基础。【方法】以B73的Su1基因组序列为模板,对170份甜玉米自交系的Su1基因进行测序,研究玉米Su1基因序列的多态性。【结果】共有97个核苷酸位点发生变异,包含85个SNP位点,12个InDel位点。其中,位于编码区的SNP位点12个,InDel位点0个;位于非编码区的SNP位点85个,InDel位点12个。编码区中SNP位点同义突变有5个,非同义突变7个。自交系材料分为76个单倍型,该基因编码区序列将自交系材料分为47种单倍型。【结论】甜玉米Su1基因经历了明显的人工选择。
中图分类号:
杨彩霞, 顾炜, 关媛, 瞿静涛, 党冬冬, 吴鹏昊, 郑洪建. 甜玉米基因Sugary1(Su1)序列的变异分析[J]. 新疆农业科学, 2024, 61(7): 1605-1614.
YANG Caixia, GU Wei, GUAN Yuan, QU Jingtao, DANG Dongdong, WU Penghao, ZHENG Hongjian. Variation analysis of sweetness gene Sugary1 (Su1) sequence in Sweet corn[J]. Xinjiang Agricultural Sciences, 2024, 61(7): 1605-1614.
| 引物 Primers | 序列(5'-3') Sequences(5'-3') | 产物长度 Product length(bp) | 退火温度 Melting temperature(℃) |
|---|---|---|---|
| 1-F | CAGAAACTGATTGCTGGGTG | 1 131 | 60 |
| 1-R | GGCAGGGCATTAGCTCTATAC | ||
| 2-F | AGCTTATAGGAAGAGATGGATAATT | 1 445 | 58 |
| 2-R | AGAGGTGTCCCTGTCGTAATC | ||
| 3-F | CAGATACTGGGTAACAGAAATGC | 1 301 | 58 |
| 3-R | GGACTGATAGATGGCTAAATGG | ||
| 4-F | CCCCTCACTGAATCCCATAAC | 1 441 | 60 |
| 4-R | CTTTTCTCTGCTCAACCCACAT | ||
| 5-F | AGGACTTCCCAGCCCAACT | 1 457 | 58 |
| 5-R | CCAGCATACCACTGACTACAAAT | ||
| 6-F | TCCGAAGATTAAGGAAGAGG | 1 467 | 60 |
| 6-R | AACTACCGACAACAGAGCAAC |
表1 Su1基因引物序列信息
Tab.1 Primer sequence information of Su1 gene
| 引物 Primers | 序列(5'-3') Sequences(5'-3') | 产物长度 Product length(bp) | 退火温度 Melting temperature(℃) |
|---|---|---|---|
| 1-F | CAGAAACTGATTGCTGGGTG | 1 131 | 60 |
| 1-R | GGCAGGGCATTAGCTCTATAC | ||
| 2-F | AGCTTATAGGAAGAGATGGATAATT | 1 445 | 58 |
| 2-R | AGAGGTGTCCCTGTCGTAATC | ||
| 3-F | CAGATACTGGGTAACAGAAATGC | 1 301 | 58 |
| 3-R | GGACTGATAGATGGCTAAATGG | ||
| 4-F | CCCCTCACTGAATCCCATAAC | 1 441 | 60 |
| 4-R | CTTTTCTCTGCTCAACCCACAT | ||
| 5-F | AGGACTTCCCAGCCCAACT | 1 457 | 58 |
| 5-R | CCAGCATACCACTGACTACAAAT | ||
| 6-F | TCCGAAGATTAAGGAAGAGG | 1 467 | 60 |
| 6-R | AACTACCGACAACAGAGCAAC |
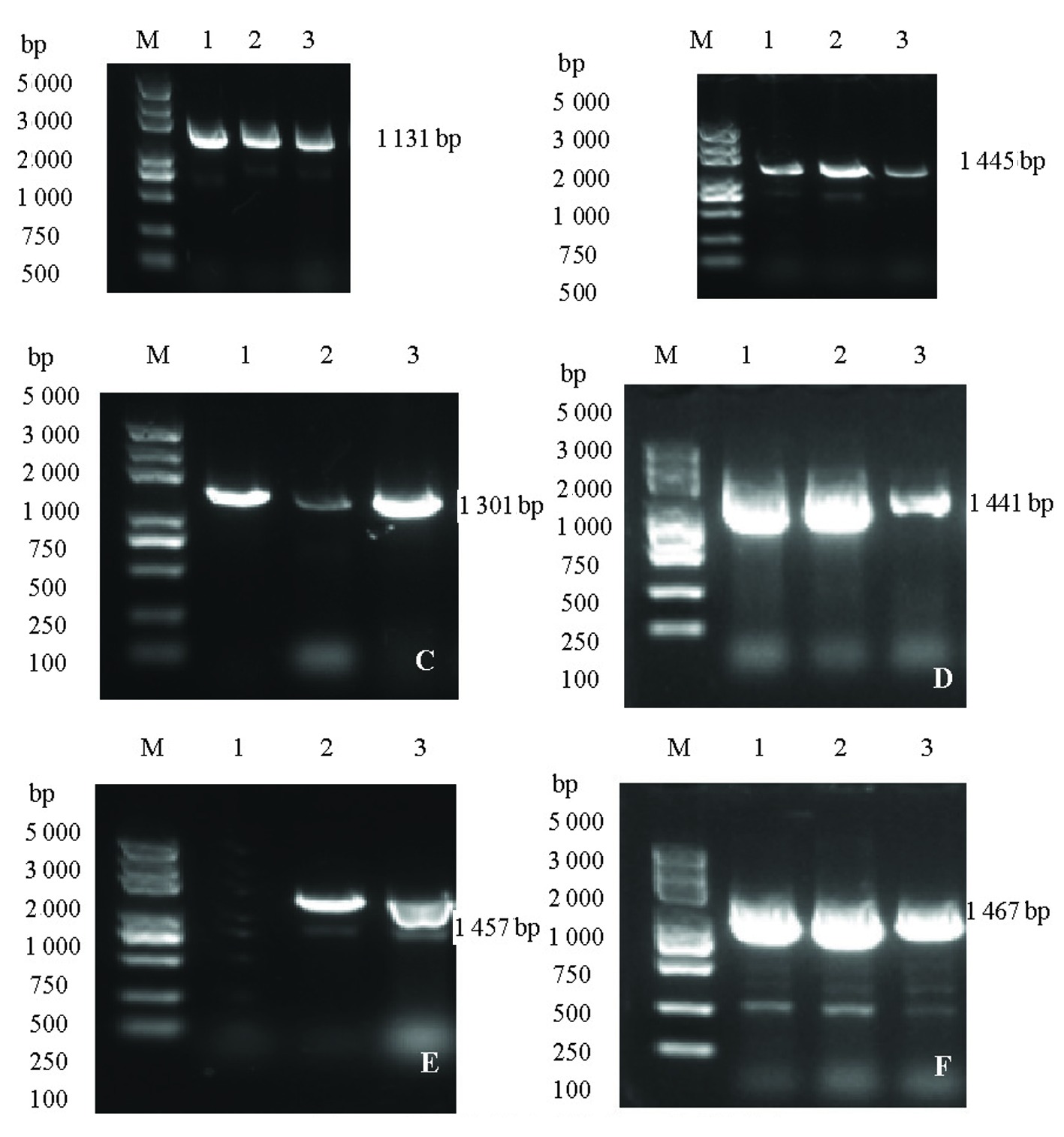
图1 甜玉米Su1基因引物检测部分材料PCR扩增电泳图 注: A:引物1-F/1-R; B:引物2-F/2-R; C:引物3-F/3-R; D:引物4-F/4-R; E:引物5-F/5-R;F:引物6-F/6-R
Fig.1 PCR amplification electrophoresis map of Su1 gene primer detection partial materials in sweet corn Note: A : primer 1-F/1-R; B : primer 2-F/2-R; C :primer 3-F/3-R; D : primer 4-F/4-R; E : Primer 5-F/5-R; F : Primer 6-F/6-R
| 位点 Site | 多态类型 Polymorphic type | 频率 Frequence | 位置 Location | 密码子变化 Coding change | 氨基酸变化 Amino Acid Mutation |
|---|---|---|---|---|---|
| InDel_261-269 | -/GTCACGA | 0.48 | intron3 | ||
| SNP_285 | G/A | 0.49 | intron3 | ||
| InDel_290 | -/T | 0.20 | intron3 | ||
| SNP_328 | A/G | 0.71 | intron | ||
| InDel_389-419 | TCTCGTTTGCAGGTTCAAATGGAGTGAAGTT /- | 0.48 | intron3 | ||
| SNP_734 | A/T | 0.22 | intron4 | ||
| SNP_746 | G/A | 0.22 | intron4 | ||
| SNP_770 | G/T | 0.49 | intron4 | ||
| SNP_863 | T/C | 0.22 | intron4 | ||
| SNP_901 | A/G | 0.71 | intron4 | ||
| InDel_930 | T/- | 0.21 | intron4 | ||
| SNP_1106 | G/C | 0.55 | exon5 | GTG/GTC | n(V/V) |
| SNP_1364 | A/G | 0.18 | intron5 | ||
| SNP_1389 | A/G | 0.19 | intron5 | ||
| SNP_1403 | T/C | 0.20 | intron5 | ||
| SNP_1572 | A/G | 0.59 | intron6 | ||
| SNP_1587 | C/T | 0.19 | intron6 | ||
| SNP_1836 | C/T | 0.19 | intron7 | ||
| SNP_1861 | A/G | 0.18 | intron7 | ||
| SNP_2054 | G/A | 0.56 | intron8 | ||
| SNP_2067 | T/G | 0.56 | intron8 | ||
| SNP_2085 | T/C | 0.36 | intron8 | ||
| SNP_2137 | T/C | 0.36 | intron8 | ||
| SNP_2498 | A/G | 0.58 | exon10 | AAT/AGT | N/S |
| SNP_2524 | A/T | 0.50 | intron10 | ||
| InDel_2542 | T/- | 0.51 | intron10 | ||
| SNP_2571 | A/G | 0.74 | intron10 | ||
| SNP_2586 | G/A | 0.24 | intron10 | ||
| SNP_2650 | C/T | 0.49 | intron10 | ||
| SNP_2713 | C/T | 0.50 | intron10 | ||
| InDel_2777 | -/AAG | 0.51 | intron10 | ||
| SNP_2880 | C/T | 0.74 | intron10 | ||
| SNP_2881 | A/G | 0.51 | intron10 | ||
| InDel_2910 | -/C | 0.24 | intron10 | ||
| SNP_3076 | T/A | 0.24 | intron10 | ||
| SNP_3107 | A/G | 0.51 | intron10 | ||
| SNP_3432 | T/C | 0.20 | intron10 | ||
| SNP_3455 | A/C | 0.74 | intron10 | ||
| InDel_3629-3635 | CTGCCA/- | 0.18 | intron10 | ||
| SNP_3636 | A/G | 0.04 | intron10 | ||
| SNP_3664 | T/C | 0.44 | intron10 | ||
| SNP_3725 | C/A | 0.45 | intron10 | ||
| SNP_3808 | T/C | 0.19 | intron10 | ||
| SNP_3856 | C/T | 0.51 | intron10 | ||
| SNP_4070 | A/G | 0.52 | intron10 | ||
| SNP_4073 | A/G | 0.54 | intron10 | ||
| A/T | 0.22 | intron10 | |||
| SNP_4153 | T/A | 0.76 | intron10 | ||
| SNP_4316 | T/C | 0.76 | intron11 | ||
| SNP_4322 | G/A | 0.52 | intron11 | ||
| SNP_4329 | G/C | 0.22 | intron11 | ||
| SNP_4386 | T/A | 0.22 | intron11 | ||
| SNP_4479 | C/T | 0.22 | intron11 | ||
| SNP_4685 | C/A | 0.21 | intron11 | ||
| SNP_4820 | A/C | 0.48 | intron11 | ||
| SNP_5037 | T/C | 0.28 | intron11 | ||
| SNP_5071 | G/A | 0.50 | intron11 | ||
| SNP_5185 | C/G | 0.74 | intron12 | ||
| SNP_5292 | A/C | 0.25 | intron12 | ||
| SNP_5369 | G/A | 0.24 | intron12 | ||
| SNP_5406 | G/T | 0.75 | intron12 | ||
| SNP_5503 | G/T | 0.50 | intron12 | ||
| SNP_5522 | G/A | 0.49 | intron12 | ||
| SNP_5758 | T/C | 0.16 | intron13 | ||
| SNP_5845 | C/A | 0.49 | intron13 | ||
| SNP_6094 | T/C | 0.75 | intron14 | ||
| SNP_6169 | T/C | 0.72 | intron14 | ||
| SNP_6283 | A/C | 0.71 | intron14 | ||
| SNP_6123 | G/A | 0.25 | intron14 | ||
| SNP_6342 | C/A | 0.72 | intron14 | ||
| SNP_6391 | C/A | 0.24 | intron14 | ||
| InDel_6449 | T/- | 0.24 | intron14 | ||
| SNP_6594 | C/A | 0.47 | intron15 | ||
| SNP_6597 | G/A | 0.24 | intron15 | ||
| SNP_6623 | A/G | 0.47 | intron15 | ||
| InDel_6628 | C/- | 0.47 | intron15 | ||
| SNP_6679 | T/C | 0.24 | exon16 | AAT/AAC | n(N/N) |
| SNP_6686 | C/T | 0.47 | exon16 | CGT/TGT | R/C |
| SNP_6721 | G/T | 0.47 | exon16 | TTG/TTT | L/F |
| SNP_6746 | A/G | 0.02 | exon16 | AAA/GAA | K/E |
| SNP_6773 | C/T | 0.47 | intron16 | ||
| SNP_6807 | C/A | 0.24 | intron16 | ||
| SNP_6870 | G/T | 0.71 | intron16 | ||
| SNP_6887 | A/G | 0.71 | exon17 | AAA/GAA | K/E |
| SNP_6956 | C/T | 0.46 | exon17 | CCC/CCT | n(P/P) |
| SNP_7053 | G/A | 0.46 | intron17 | ||
| InDel_7083 | G/- | 0.23 | intron17 | ||
| InDel_7084 | A/- | 0.23 | intron17 | ||
| SNP_7086 | C/T | 0.46 | intron17 | ||
| SNP_7087 | G/A | 0.23 | intron17 | ||
| SNP_7192 | T/C | 0.69 | exon18 | GTT/GTC | n(V/V) |
| SNP_7218 | G/A | 0.46 | exon18 | CGA/CAA | R/Q |
| SNP_7279 | G/A | 0.17 | exon18 | CTG/CTA | n(L/L) |
| SNP_7379 | G/C | 0.46 | exon18 | GAT/CAT | D/H |
| SNP_7395 | A/G | 0.69 | 3'UTR | ||
| SNP_7397 | A/T | 0.46 | 3'UTR | ||
| SNP_7414 | A/G | 0.69 | 3'UTR |
表2 甜玉米Su1基因编码区多态性
Tab.2 Polymorphism of Su1 gene coding region in sweet corn
| 位点 Site | 多态类型 Polymorphic type | 频率 Frequence | 位置 Location | 密码子变化 Coding change | 氨基酸变化 Amino Acid Mutation |
|---|---|---|---|---|---|
| InDel_261-269 | -/GTCACGA | 0.48 | intron3 | ||
| SNP_285 | G/A | 0.49 | intron3 | ||
| InDel_290 | -/T | 0.20 | intron3 | ||
| SNP_328 | A/G | 0.71 | intron | ||
| InDel_389-419 | TCTCGTTTGCAGGTTCAAATGGAGTGAAGTT /- | 0.48 | intron3 | ||
| SNP_734 | A/T | 0.22 | intron4 | ||
| SNP_746 | G/A | 0.22 | intron4 | ||
| SNP_770 | G/T | 0.49 | intron4 | ||
| SNP_863 | T/C | 0.22 | intron4 | ||
| SNP_901 | A/G | 0.71 | intron4 | ||
| InDel_930 | T/- | 0.21 | intron4 | ||
| SNP_1106 | G/C | 0.55 | exon5 | GTG/GTC | n(V/V) |
| SNP_1364 | A/G | 0.18 | intron5 | ||
| SNP_1389 | A/G | 0.19 | intron5 | ||
| SNP_1403 | T/C | 0.20 | intron5 | ||
| SNP_1572 | A/G | 0.59 | intron6 | ||
| SNP_1587 | C/T | 0.19 | intron6 | ||
| SNP_1836 | C/T | 0.19 | intron7 | ||
| SNP_1861 | A/G | 0.18 | intron7 | ||
| SNP_2054 | G/A | 0.56 | intron8 | ||
| SNP_2067 | T/G | 0.56 | intron8 | ||
| SNP_2085 | T/C | 0.36 | intron8 | ||
| SNP_2137 | T/C | 0.36 | intron8 | ||
| SNP_2498 | A/G | 0.58 | exon10 | AAT/AGT | N/S |
| SNP_2524 | A/T | 0.50 | intron10 | ||
| InDel_2542 | T/- | 0.51 | intron10 | ||
| SNP_2571 | A/G | 0.74 | intron10 | ||
| SNP_2586 | G/A | 0.24 | intron10 | ||
| SNP_2650 | C/T | 0.49 | intron10 | ||
| SNP_2713 | C/T | 0.50 | intron10 | ||
| InDel_2777 | -/AAG | 0.51 | intron10 | ||
| SNP_2880 | C/T | 0.74 | intron10 | ||
| SNP_2881 | A/G | 0.51 | intron10 | ||
| InDel_2910 | -/C | 0.24 | intron10 | ||
| SNP_3076 | T/A | 0.24 | intron10 | ||
| SNP_3107 | A/G | 0.51 | intron10 | ||
| SNP_3432 | T/C | 0.20 | intron10 | ||
| SNP_3455 | A/C | 0.74 | intron10 | ||
| InDel_3629-3635 | CTGCCA/- | 0.18 | intron10 | ||
| SNP_3636 | A/G | 0.04 | intron10 | ||
| SNP_3664 | T/C | 0.44 | intron10 | ||
| SNP_3725 | C/A | 0.45 | intron10 | ||
| SNP_3808 | T/C | 0.19 | intron10 | ||
| SNP_3856 | C/T | 0.51 | intron10 | ||
| SNP_4070 | A/G | 0.52 | intron10 | ||
| SNP_4073 | A/G | 0.54 | intron10 | ||
| A/T | 0.22 | intron10 | |||
| SNP_4153 | T/A | 0.76 | intron10 | ||
| SNP_4316 | T/C | 0.76 | intron11 | ||
| SNP_4322 | G/A | 0.52 | intron11 | ||
| SNP_4329 | G/C | 0.22 | intron11 | ||
| SNP_4386 | T/A | 0.22 | intron11 | ||
| SNP_4479 | C/T | 0.22 | intron11 | ||
| SNP_4685 | C/A | 0.21 | intron11 | ||
| SNP_4820 | A/C | 0.48 | intron11 | ||
| SNP_5037 | T/C | 0.28 | intron11 | ||
| SNP_5071 | G/A | 0.50 | intron11 | ||
| SNP_5185 | C/G | 0.74 | intron12 | ||
| SNP_5292 | A/C | 0.25 | intron12 | ||
| SNP_5369 | G/A | 0.24 | intron12 | ||
| SNP_5406 | G/T | 0.75 | intron12 | ||
| SNP_5503 | G/T | 0.50 | intron12 | ||
| SNP_5522 | G/A | 0.49 | intron12 | ||
| SNP_5758 | T/C | 0.16 | intron13 | ||
| SNP_5845 | C/A | 0.49 | intron13 | ||
| SNP_6094 | T/C | 0.75 | intron14 | ||
| SNP_6169 | T/C | 0.72 | intron14 | ||
| SNP_6283 | A/C | 0.71 | intron14 | ||
| SNP_6123 | G/A | 0.25 | intron14 | ||
| SNP_6342 | C/A | 0.72 | intron14 | ||
| SNP_6391 | C/A | 0.24 | intron14 | ||
| InDel_6449 | T/- | 0.24 | intron14 | ||
| SNP_6594 | C/A | 0.47 | intron15 | ||
| SNP_6597 | G/A | 0.24 | intron15 | ||
| SNP_6623 | A/G | 0.47 | intron15 | ||
| InDel_6628 | C/- | 0.47 | intron15 | ||
| SNP_6679 | T/C | 0.24 | exon16 | AAT/AAC | n(N/N) |
| SNP_6686 | C/T | 0.47 | exon16 | CGT/TGT | R/C |
| SNP_6721 | G/T | 0.47 | exon16 | TTG/TTT | L/F |
| SNP_6746 | A/G | 0.02 | exon16 | AAA/GAA | K/E |
| SNP_6773 | C/T | 0.47 | intron16 | ||
| SNP_6807 | C/A | 0.24 | intron16 | ||
| SNP_6870 | G/T | 0.71 | intron16 | ||
| SNP_6887 | A/G | 0.71 | exon17 | AAA/GAA | K/E |
| SNP_6956 | C/T | 0.46 | exon17 | CCC/CCT | n(P/P) |
| SNP_7053 | G/A | 0.46 | intron17 | ||
| InDel_7083 | G/- | 0.23 | intron17 | ||
| InDel_7084 | A/- | 0.23 | intron17 | ||
| SNP_7086 | C/T | 0.46 | intron17 | ||
| SNP_7087 | G/A | 0.23 | intron17 | ||
| SNP_7192 | T/C | 0.69 | exon18 | GTT/GTC | n(V/V) |
| SNP_7218 | G/A | 0.46 | exon18 | CGA/CAA | R/Q |
| SNP_7279 | G/A | 0.17 | exon18 | CTG/CTA | n(L/L) |
| SNP_7379 | G/C | 0.46 | exon18 | GAT/CAT | D/H |
| SNP_7395 | A/G | 0.69 | 3'UTR | ||
| SNP_7397 | A/T | 0.46 | 3'UTR | ||
| SNP_7414 | A/G | 0.69 | 3'UTR |
| 参数 Parameter | 序列长度 Product length (bp) | SNPs+InDel 数目 Number of SNP+Indel | SNP+Indel 变异频率 Frequency of SNP+Indel (bp) | SNP 数目 Number of SNP | SNP 频率 Frequency of SNP (bp) | InDels 数目 Number of Indel | Indel 频率 Frequency of Indel (bp) |
|---|---|---|---|---|---|---|---|
| 全长 | 7 438 | 161 | 0.022 | 86 | 0.012 | 75 | 0.010 |
| intron3 | 331 | 41 | 0.124 | 2 | 0.006 | 39 | 0.118 |
| exon4 | 154 | 0 | 0 | 0 | 0 | 0 | 0 |
| intron4 | 456 | 7 | 0.015 | 6 | 0.013 | 1 | 0.002 |
| exon5 | 77 | 1 | 0.013 | 1 | 0.013 | 0 | 0 |
| intron5 | 247 | 3 | 0.012 | 3 | 0.012 | 0 | 0 |
| exon6 | 164 | 0 | 0 | 0 | 0 | 0 | 0 |
| intron6 | 91 | 2 | 0.022 | 2 | 0.022 | 0 | 0 |
| exon7 | 107 | 0 | 0 | 0 | 0 | 0 | 0 |
| intron7 | 118 | 2 | 0.017 | 2 | 0.017 | 0 | 0 |
| exon8 | 85 | 0 | 0 | 0 | 0 | 0 | 0 |
| intron8 | 270 | 4 | 0.015 | 4 | 0.015 | 0 | 0 |
| exon9 | 72 | 0 | 0 | 0 | 0 | 0 | 0 |
| intron9 | 73 | 0 | 0 | 0 | 0 | 0 | 0 |
| exon10 | 127 | 1 | 0.008 | 1 | 0.008 | 0 | 0 |
| intron10 | 1716 | 32 | 0.019 | 19 | 0.011 | 13 | 0.008 |
| exon11 | 87 | 0 | 0 | 0 | 0 | 0 | 0 |
| intron11 | 776 | 26 | 0.034 | 9 | 0.012 | 17 | 0.022 |
| exon12 | 94 | 0 | 0 | 0 | 0 | 0 | 0 |
| intron12 | 437 | 6 | 0.014 | 6 | 0.014 | 0 | 0 |
| exon13 | 159 | 1 | 0.006 | 1 | 0.006 | 0 | 0 |
| intron13 | 223 | 1 | 0.004 | 1 | 0.004 | 0 | 0 |
| exon14 | 84 | 0 | 0 | 0 | 0 | 0 | 0 |
| intron14 | 421 | 7 | 0.017 | 6 | 0.014 | 1 | 0.002 |
| exon15 | 84 | 0 | 0 | 0 | 0 | 0 | 0 |
| intron15 | 93 | 4 | 0.043 | 3 | 0.032 | 1 | 0.011 |
| exon16 | 79 | 4 | 0.051 | 4 | 0.051 | 0 | 0 |
| intron16 | 130 | 3 | 0.023 | 3 | 0.023 | 0 | 0 |
| exon17 | 118 | 2 | 0.017 | 2 | 0.017 | 0 | 0 |
| intron17 | 125 | 6 | 0.048 | 4 | 0.032 | 2 | 0.016 |
| exon18 | 258 | 4 | 0.016 | 4 | 0.016 | 0 | 0 |
表3 甜玉米Su1基因序列变异参数
Tab.3 Parameters for the sequence of sweet corn Su1 gene
| 参数 Parameter | 序列长度 Product length (bp) | SNPs+InDel 数目 Number of SNP+Indel | SNP+Indel 变异频率 Frequency of SNP+Indel (bp) | SNP 数目 Number of SNP | SNP 频率 Frequency of SNP (bp) | InDels 数目 Number of Indel | Indel 频率 Frequency of Indel (bp) |
|---|---|---|---|---|---|---|---|
| 全长 | 7 438 | 161 | 0.022 | 86 | 0.012 | 75 | 0.010 |
| intron3 | 331 | 41 | 0.124 | 2 | 0.006 | 39 | 0.118 |
| exon4 | 154 | 0 | 0 | 0 | 0 | 0 | 0 |
| intron4 | 456 | 7 | 0.015 | 6 | 0.013 | 1 | 0.002 |
| exon5 | 77 | 1 | 0.013 | 1 | 0.013 | 0 | 0 |
| intron5 | 247 | 3 | 0.012 | 3 | 0.012 | 0 | 0 |
| exon6 | 164 | 0 | 0 | 0 | 0 | 0 | 0 |
| intron6 | 91 | 2 | 0.022 | 2 | 0.022 | 0 | 0 |
| exon7 | 107 | 0 | 0 | 0 | 0 | 0 | 0 |
| intron7 | 118 | 2 | 0.017 | 2 | 0.017 | 0 | 0 |
| exon8 | 85 | 0 | 0 | 0 | 0 | 0 | 0 |
| intron8 | 270 | 4 | 0.015 | 4 | 0.015 | 0 | 0 |
| exon9 | 72 | 0 | 0 | 0 | 0 | 0 | 0 |
| intron9 | 73 | 0 | 0 | 0 | 0 | 0 | 0 |
| exon10 | 127 | 1 | 0.008 | 1 | 0.008 | 0 | 0 |
| intron10 | 1716 | 32 | 0.019 | 19 | 0.011 | 13 | 0.008 |
| exon11 | 87 | 0 | 0 | 0 | 0 | 0 | 0 |
| intron11 | 776 | 26 | 0.034 | 9 | 0.012 | 17 | 0.022 |
| exon12 | 94 | 0 | 0 | 0 | 0 | 0 | 0 |
| intron12 | 437 | 6 | 0.014 | 6 | 0.014 | 0 | 0 |
| exon13 | 159 | 1 | 0.006 | 1 | 0.006 | 0 | 0 |
| intron13 | 223 | 1 | 0.004 | 1 | 0.004 | 0 | 0 |
| exon14 | 84 | 0 | 0 | 0 | 0 | 0 | 0 |
| intron14 | 421 | 7 | 0.017 | 6 | 0.014 | 1 | 0.002 |
| exon15 | 84 | 0 | 0 | 0 | 0 | 0 | 0 |
| intron15 | 93 | 4 | 0.043 | 3 | 0.032 | 1 | 0.011 |
| exon16 | 79 | 4 | 0.051 | 4 | 0.051 | 0 | 0 |
| intron16 | 130 | 3 | 0.023 | 3 | 0.023 | 0 | 0 |
| exon17 | 118 | 2 | 0.017 | 2 | 0.017 | 0 | 0 |
| intron17 | 125 | 6 | 0.048 | 4 | 0.032 | 2 | 0.016 |
| exon18 | 258 | 4 | 0.016 | 4 | 0.016 | 0 | 0 |
| π | θ | Tajima’s D | Fu and Li’s D* | Fu and Li’s F* | |||
|---|---|---|---|---|---|---|---|
| 全长 | 0.004 81 | 0.002 7 | 4.140 3 | 2.408 2 | 3.829 | ||
| intron3 | 0.003 15 | 0.001 2 | 2.275 31* | 0.646 81 | 1.367 3 | ||
| exon4 | 0 | 0 | 0 | 0 | 0 | ||
| intron4 | 0.004 31 | 0.002 31 | 1.796 59* | 0.569 9 | 0.774 04 | ||
| exon5 | 0.006 47 | 0.002 27 | 0 | 0 | 0 | ||
| intron5 | 0.003 77 | 0.002 12 | 1.273 95 | 0.784 19 | 1.112 72 | ||
| exon6 | 0 | 0 | 0 | 0 | 0 | ||
| intron6 | 0.010 48 | 0.003 85 | 2.414 01 | 0.646 81 | 1.418 9 | ||
| exon7 | 0 | 0 | 0 | 0 | 0 | ||
| intron7 | 0.005 19 | 0.002 97 | 1.047 98 | 0.646 81 | 0.910 65 | ||
| exon8 | 0 | 0 | 0 | 0 | 0 | ||
| intron8 | 0.007 12 | 0.002 59 | 3.178 3 | 0.896 55 | 1.952 19* | ||
| exon9 | 0 | 0 | 0 | 0 | 0 | ||
| intron9 | 0 | 0 | 0 | 0 | 0 | ||
| exon10 | 0.003 96 | 0.001 38 | 1.945 52 | 0.462 13 | 1.078 16 | ||
| intron10 | 0.004 78 | 0.002 05 | 3.603 8 | 1.748 2 | 2.970 4 | ||
| exon11 | 0 | 0 | 0 | 0 | 0 | ||
| intron11 | 0.004 81 | 0.002 07 | 3.058 0 | 1.283 1 | 2.284 9 | ||
| exon12 | 0 | 0 | 0 | 0 | 0 | ||
| intron12 | 0.005 75 | 0.002 4 | 2.893 4 | 1.077 5 | 2.017 07* | ||
| exon13 | 0 | 0 | 0 | 0 | 0 | ||
| intron13 | 0.002 25 | 0.000 78 | 1.945 52 | 0.462 13 | 1.078 16 | ||
| exon14 | 0 | 0 | 0 | 0 | 0 | ||
| intron14 | 0.005 6 | 0.002 5 | 2.572 35* | 1.077 05 | 1.886 68 | ||
| exon15 | 0 | 0 | 0 | 0 | 0 | ||
| intron15 | 0.014 81 | 0.005 7 | 2.617 73* | 0.784 19 | 1.625 72 | ||
| exon16 | 0.017 9 | 0.008 86 | 1.858 31 | 0.896 55 | 1.436 61 | ||
| intron16 | 0.009 91 | 0.004 04 | 2.387 60* | 0.784 19 | 1.537 87 | ||
| exon17 | 0.007 81 | 0.002 97 | 2.285 467* | 0.646 81 | 1.370 78 | ||
| intron17 | 0.013 35 | 0.005 69 | 2.450 52* | 0.896 55 | 1.667 92 | ||
表4 中性检验相关参数
Tab.4 Neutral test related parameters
| π | θ | Tajima’s D | Fu and Li’s D* | Fu and Li’s F* | |||
|---|---|---|---|---|---|---|---|
| 全长 | 0.004 81 | 0.002 7 | 4.140 3 | 2.408 2 | 3.829 | ||
| intron3 | 0.003 15 | 0.001 2 | 2.275 31* | 0.646 81 | 1.367 3 | ||
| exon4 | 0 | 0 | 0 | 0 | 0 | ||
| intron4 | 0.004 31 | 0.002 31 | 1.796 59* | 0.569 9 | 0.774 04 | ||
| exon5 | 0.006 47 | 0.002 27 | 0 | 0 | 0 | ||
| intron5 | 0.003 77 | 0.002 12 | 1.273 95 | 0.784 19 | 1.112 72 | ||
| exon6 | 0 | 0 | 0 | 0 | 0 | ||
| intron6 | 0.010 48 | 0.003 85 | 2.414 01 | 0.646 81 | 1.418 9 | ||
| exon7 | 0 | 0 | 0 | 0 | 0 | ||
| intron7 | 0.005 19 | 0.002 97 | 1.047 98 | 0.646 81 | 0.910 65 | ||
| exon8 | 0 | 0 | 0 | 0 | 0 | ||
| intron8 | 0.007 12 | 0.002 59 | 3.178 3 | 0.896 55 | 1.952 19* | ||
| exon9 | 0 | 0 | 0 | 0 | 0 | ||
| intron9 | 0 | 0 | 0 | 0 | 0 | ||
| exon10 | 0.003 96 | 0.001 38 | 1.945 52 | 0.462 13 | 1.078 16 | ||
| intron10 | 0.004 78 | 0.002 05 | 3.603 8 | 1.748 2 | 2.970 4 | ||
| exon11 | 0 | 0 | 0 | 0 | 0 | ||
| intron11 | 0.004 81 | 0.002 07 | 3.058 0 | 1.283 1 | 2.284 9 | ||
| exon12 | 0 | 0 | 0 | 0 | 0 | ||
| intron12 | 0.005 75 | 0.002 4 | 2.893 4 | 1.077 5 | 2.017 07* | ||
| exon13 | 0 | 0 | 0 | 0 | 0 | ||
| intron13 | 0.002 25 | 0.000 78 | 1.945 52 | 0.462 13 | 1.078 16 | ||
| exon14 | 0 | 0 | 0 | 0 | 0 | ||
| intron14 | 0.005 6 | 0.002 5 | 2.572 35* | 1.077 05 | 1.886 68 | ||
| exon15 | 0 | 0 | 0 | 0 | 0 | ||
| intron15 | 0.014 81 | 0.005 7 | 2.617 73* | 0.784 19 | 1.625 72 | ||
| exon16 | 0.017 9 | 0.008 86 | 1.858 31 | 0.896 55 | 1.436 61 | ||
| intron16 | 0.009 91 | 0.004 04 | 2.387 60* | 0.784 19 | 1.537 87 | ||
| exon17 | 0.007 81 | 0.002 97 | 2.285 467* | 0.646 81 | 1.370 78 | ||
| intron17 | 0.013 35 | 0.005 69 | 2.450 52* | 0.896 55 | 1.667 92 | ||
| [1] | 陈骁熠, 瞿宏杰. 甜玉米的品质改良技术[J]. 农村经济与科技, 2001, 12(8): 15. |
| CHEN Xiaoyi, QU Hongjie. Quality improvement technology of sweet corn[J]. Rural Economy and Science-Technology, 2001, 12(8): 15. | |
| [2] | 曹忠良, 张俊江. 甜玉米的类型及关键性技术[J]. 民营科技, 2013, (5): 212. |
| CAO Zhongliang, ZHANG Junjiang. Types of sweet corn and key technologies[J]. NonState Running Science Technology Enterprises, 2013, (5): 212. | |
| [3] | 蒙云飞, 冯云敢, 贺囡囡, 等. 甜玉米的起源和分布及遗传基因研究进展[J]. 现代农业科技, 2017, (5): 4-5. |
| MENG Yunfei, FENG Yungan, HE Nannan, et al. Research progress on origin distribution and genetic genes of sweet corn[J]. Modern Agricultural Science and Technology, 2017, (5): 4-5. | |
| [4] | 邵振中. 甜玉米的研究现状与发展[J]. 襄樊职业技术学院学报, 2004, 3(4): 23-25. |
| SHAO Zhenzhong. The current situation of research and development of sweet corn[J]. Journal of Xiangfan Vocational and Technical College, 2004, 3(4): 23-25. | |
| [5] | Zhang X, Mogel K J H V, Lor V S, et al. Maizesugary enhancer 1 (se1) is a gene affecting endosperm starch metabolism[J]. Proceedings of the National Academy of Sciences of the United States of America, 2019, 116(41): 20776-20785. |
| [6] | Ferguson J E, Dickinson D B, Rhodes A M. Analysis of endosperm sugars in a sweet corn inbred (illinois 677a) which contains the sugary enhancer (se) gene and comparison of se with other corn genotypes[J]. Plant Physiology, 1979, 63(3): 416-420. |
| [7] | Chhabra R, Muthusamy V, Gain N, et al. Allelic variation in sugary1 gene affecting kernel sweetness among diverse-mutant and-wild-type maize inbreds[J]. Molecular Genetics and Genomics: MGG, 2021, 296(5): 1085-1102. |
| [8] | La Bonte D R, Juvik J A. Characterization ofsugary-1 (su-1) sugary enhancer (se) Kernels in Segregating Sweet Corn Populations[J]. Journal of the American Society for Horticultural Science, 1990, 115(1): 153-157. |
| [9] | Schultz J A, Juvik J A. Current models for starch synthesis and the sugary enhancer1 (se1) mutation inZea mays[J]. Plant Physiology and Biochemistry: PPB, 2004, 42(6): 457-464. |
| [10] | Yu J K, Moon Y S. Corn starch: quality and quantity improvement for industrial uses[J]. Plants, 2021, 11(1): 92. |
| [11] | Pan D, Nelson O E. A debranching enzyme deficiency in endosperms of the sugary-1 mutants of maize[J]. Plant Physiology, 1984, 74(2): 324-328. |
| [12] | Trimble L, Shuler S, Tracy W F. Characterization of five naturally occurring alleles at the Sugary1 locus for seed composition, seedling emergence, and Isoamylase1 activity[J]. Crop Science, 2016, 56(4): 1927-1939. |
| [13] | Rahman A, et al. Characterization of SU1 isoamylase, a determinant of storage starch structure in maize[J]. Plant Physiology, 1998, 117(2): 425-435. |
| [14] | Revilla P, Anibas C M, Tracy W F. Sweet Corn Research around the World 2015-2020[J]. Agronomy, 2021, 11(3): 534. |
| [15] | Szöke L, Moloi M J, Kovács G E, et al. The application of phytohormones as biostimulants in corn smut infected Hungarian sweet and fodder corn hybrids[J]. Plants, 2021, 10(9): 1822. |
| [16] | Chourey P S, Nelson O E. The enzymatic deficiency conditioned by the shrunken-1 mutations in maize[J]. Biochemical Genetics, 1976, 14(11/12): 1041-1055. |
| [17] | Trimble L, Shuler S, Tracy W F. Characterization of Five NaturallyOccurring Alleles at theSugary1Locus for Seed Composition, Seedling Emergence, andIsoamylase1 Activity[J]. Crop Science, 2016, 56(4):1927-1939. |
| [18] | 顾炜. 玉米籽粒突变基因Mn219和Dek39的图位克隆与基因功能分析[D]. 北京: 中国农业大学. 2018. |
| GU Wei. The map-based cloning and functional analysis of MN219 and Dek39 in maize[D]. Beijing: China Agricultural University, 2018. | |
| [19] | Rozas J, Ferrer-Mata A, Sánchez-DelBarrio J C, et al. DnaSP 6: DNA sequence polymorphism analysis of large data sets[J]. Molecular Biology and Evolution, 2017, 34(12): 3299-3302. |
| [20] | Fu Y X, Li W H. Statistical tests of neutrality of mutations[J]. Genetics, 1993, 133(3): 693-709. |
| [21] | Tajima F. Statistical method for testing the neutral mutation hypothesis by DNA polymorphism[J]. Genetics, 1989, 123(3): 585-595. |
| [22] | 杨泽峰, 张恩盈, 蒋莹, 等. 玉米耐旱相关基因dbf1的序列变异分析[J]. 玉米科学, 2014, 22(5): 20-28. |
| YANG Zefeng, ZHANG Enying, JIANG Ying, et al. Nucleotide diversity of drought tolerance related maize gene dbf1[J]. Journal of Maize Sciences, 2014, 22(5): 20-28. | |
| [23] | Ching A, Caldwell K S, Jung M, et al. SNP frequency, haplotype structure and linkage disequilibrium in elite maize inbred lines[J]. BMC Genetics, 2002, 3: 19. |
| [1] | 钟辉丽, 武均, 陆祥生. 甜玉米不同生育期施用改良剂组合对其产量及河西走廊次生盐碱化土壤性质的影响[J]. 新疆农业科学, 2024, 61(7): 1615-1625. |
| [2] | 王凯迪, 高晨旭, 裴文锋, 杨书贤, 张文庆, 宋吉坤, 马建江, 王莉, 于霁雯, 陈全家. 陆地棉TRM基因家族的鉴定及纤维品质相关优异单倍型分析[J]. 新疆农业科学, 2024, 61(3): 521-536. |
| [3] | 王丽丽, 战帅帅, 谢磊, 王继庆, 哈尼开·马坎, 任毅, 时佳, 耿洪伟. 新疆小麦籽粒过氧化物酶(POD)活性检测及其基因等位变异检测[J]. 新疆农业科学, 2020, 57(10): 1765-1774. |
| [4] | 哈力旦·依克热木, 周安定, 芦静, 曹俊梅, 刘联正, 张新忠. 新疆部分小麦材料淀粉品质基因等位变异的分子鉴定及其分布[J]. 新疆农业科学, 2017, 54(12): 2157-2163. |
| [5] | 哈力旦·依克热木;芦静;周安定;曾潮武;曹俊梅;梁晓东;刘联正;范贵强;张新忠. 新疆部分小麦材料籽粒硬度基因等位变异的分子鉴定及其分布[J]. , 2016, 53(6): 981-986. |
| [6] | 王宏飞;丛花;章艳凤;严勇亮;尤明山;倪中福. 新疆冬小麦地方品种高分子量谷蛋白亚基等位变异及其品质分析[J]. , 2011, 48(8): 1392-1398. |
| [7] | 丛花;王宏飞;章艳凤;严勇亮;池田达哉;高田兼则;长峰司. 新疆小麦地方品种籽粒硬度及其Puroindoline 基因等位变异研究[J]. , 2011, 48(6): 1056-1063. |
| [8] | 桑志勤;陈树宾;王友德;王婷;郭斌;段震宇;李玉梅. 甜玉米产量及相关农艺因素的关联分析[J]. , 2010, 47(10): 1939-1942. |
| [9] | 李进;赵红娟;梁晓玲;阿布来提·阿布拉;李铭东;雷志刚;阿依古丽·艾买提;韩登旭;邵红雨. 早熟水果型超甜玉米新特玉5号的选育研究[J]. , 2008, 45(5): 976-979. |
| [10] | 杨惠. 鲜食加甜玉米新甜玉5号的选育与应用[J]. , 2007, 44(z2): 89-91. |
| [11] | 王友德;陈树宾;王婷. 国内甜、糯玉米品种引进种植效果初报[J]. , 2002, 39(6): 341-342. |
| 阅读次数 | ||||||||||||||||||||||||||||||||||||||||||||||||||
|
全文 36
|
|
|||||||||||||||||||||||||||||||||||||||||||||||||
|
摘要 149
|
|
|||||||||||||||||||||||||||||||||||||||||||||||||